Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines
- PMID: 26983574
- PMCID: PMC4794844
- DOI: 10.1186/s12885-016-2273-6
Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines
Abstract
Background: Post-transcriptional regulation by microRNAs is recognized as one of the major pathways for the control of cellular homeostasis. Less well understood is the transcriptional and epigenetic regulation of genes encoding microRNAs. In the present study we addressed the epigenetic regulation of the miR-181c in normal and malignant brain cells.
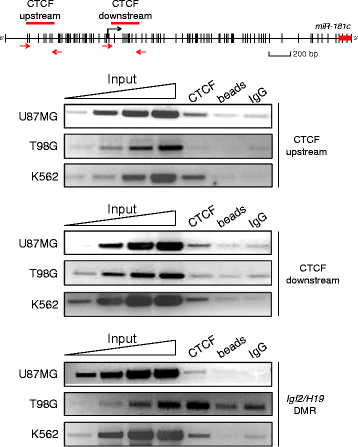
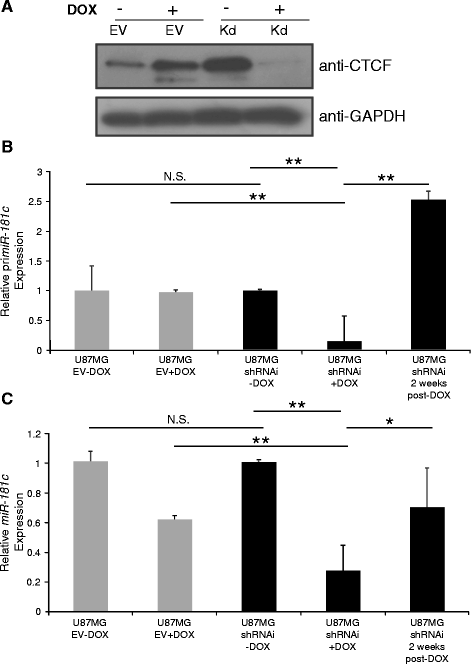
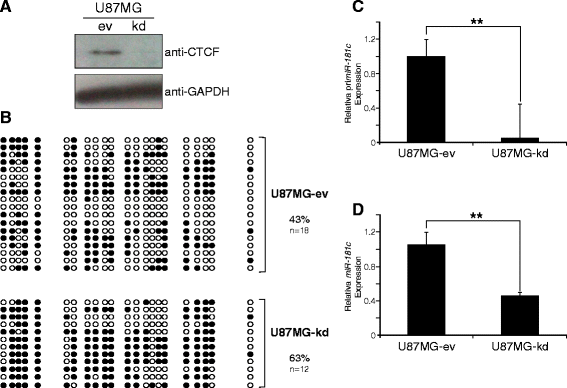
Methods: To explore the epigenetic regulation of the miR-181c we evaluated its expression using RT-qPCR and the in vivo binding of the CCCTC-binding factor (CTCF) to its regulatory region in different glioblastoma cell lines. DNA methylation survey, chromatin immunoprecipitation and RNA interference assays were used to assess the role of CTCF in the miR-181c epigenetic silencing.
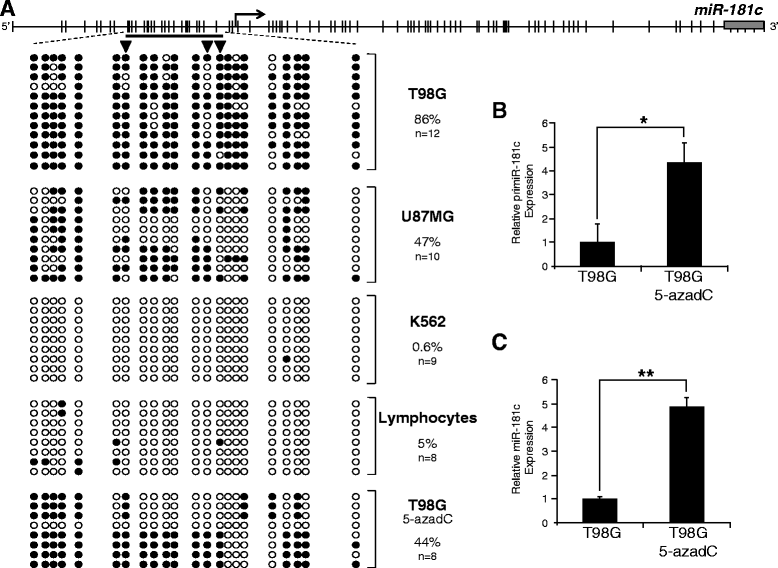
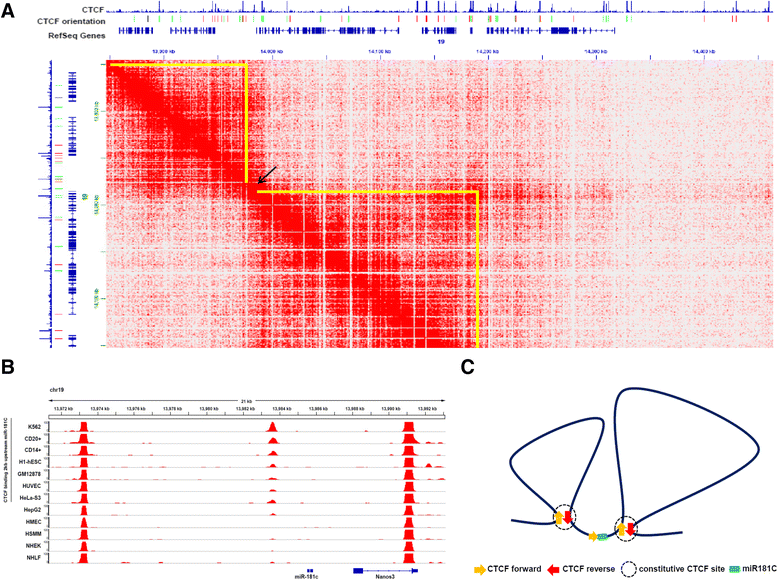
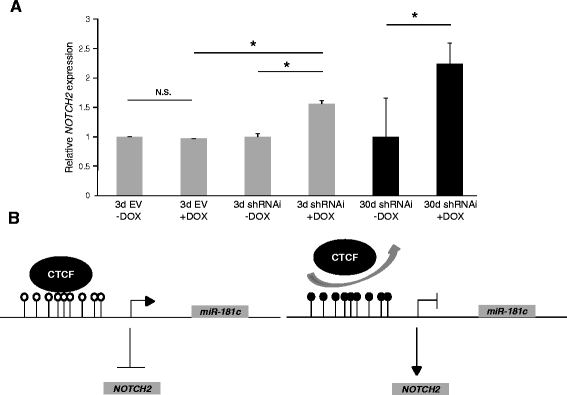
Results: We found that miR-181c is downregulated in glioblastoma cell lines, as compared to normal brain tissues. Loss of expression correlated with a notorious gain of DNA methylation at the miR-181c promoter region and the dissociation of the multifunctional nuclear factor CTCF. Taking advantage of the genomic distribution of CTCF in different cell types we propose that CTCF has a local and cell type specific regulatory role over the miR-181c and not an architectural one through chromatin loop formation. This is supported by the depletion of CTCF in glioblastoma cells affecting the expression levels of NOTCH2 as a target of miR-181c.
Conclusion: Together, our results point to the epigenetic role of CTCF in the regulation of microRNAs implicated in tumorigenesis.
Keywords: CCCTC-binding factor (CTCF); DNA methylation; Epigenetics; Glioblastoma cells; RNA interference.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

