The Capacity to Act in Trans Varies Among Drosophila Enhancers
- PMID: 26984057
- PMCID: PMC4858774
- DOI: 10.1534/genetics.115.185645
The Capacity to Act in Trans Varies Among Drosophila Enhancers
Abstract
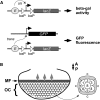
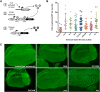
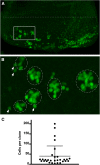
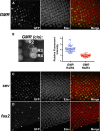
The interphase nucleus is organized such that genomic segments interact in cis, on the same chromosome, and in trans, between different chromosomes. In Drosophila and other Dipterans, extensive interactions are observed between homologous chromosomes, which can permit enhancers and promoters to communicate in trans Enhancer action in trans has been observed for a handful of genes in Drosophila, but it is as yet unclear whether this is a general property of all enhancers or specific to a few. Here, we test a collection of well-characterized enhancers for the capacity to act in trans Specifically, we tested 18 enhancers that are active in either the eye or wing disc of third instar Drosophila larvae and, using two different assays, found evidence that each enhancer can act in trans However, the degree to which trans-action was supported varied greatly between enhancers. Quantitative analysis of enhancer activity supports a model wherein an enhancer's strength of transcriptional activation is a major determinant of its ability to act in trans, but that additional factors may also contribute to an enhancer's trans-activity. In sum, our data suggest that a capacity to activate a promoter on a paired chromosome is common among Drosophila enhancers.
Keywords: RMCE; interchromosomal interactions; long-range enhancer; somatic homolog pairing; transvection.
Copyright © 2016 by the Genetics Society of America.
Figures




References
-
- Apostolou E., Thanos D., 2008. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134: 85–96. - PubMed
-
- Bacher C. P., Guggiari M., Brors B., Augui S., Clerc P., et al. , 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8: 293–299. - PubMed
-
- Bantignies F., Cavalli G., 2011. Polycomb group proteins: repression in 3D. Trends Genet. 27: 454–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

