DASPfind: new efficient method to predict drug-target interactions
- PMID: 26985240
- PMCID: PMC4793623
- DOI: 10.1186/s13321-016-0128-4
DASPfind: new efficient method to predict drug-target interactions
Abstract
Background: Identification of novel drug-target interactions (DTIs) is important for drug discovery. Experimental determination of such DTIs is costly and time consuming, hence it necessitates the development of efficient computational methods for the accurate prediction of potential DTIs. To-date, many computational methods have been proposed for this purpose, but they suffer the drawback of a high rate of false positive predictions.
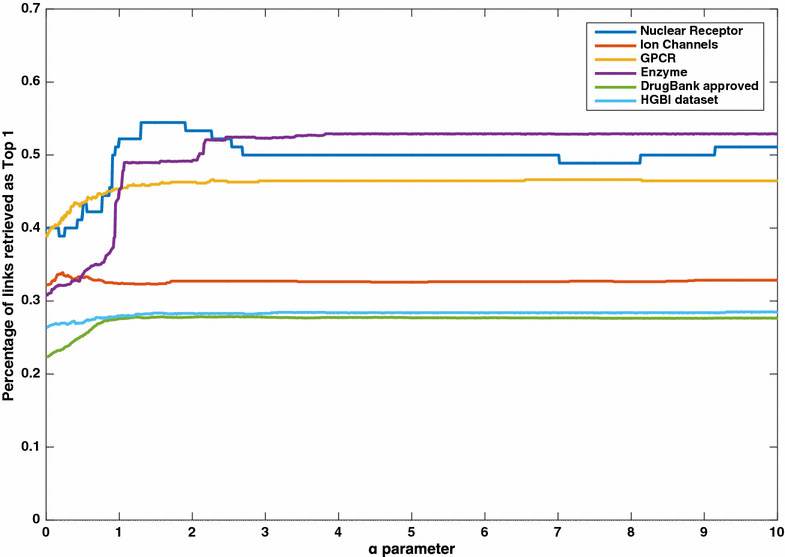
Results: Here, we developed a novel computational DTI prediction method, DASPfind. DASPfind uses simple paths of particular lengths inferred from a graph that describes DTIs, similarities between drugs, and similarities between the protein targets of drugs. We show that on average, over the four gold standard DTI datasets, DASPfind significantly outperforms other existing methods when the single top-ranked predictions are considered, resulting in 46.17 % of these predictions being correct, and it achieves 49.22 % correct single top ranked predictions when the set of all DTIs for a single drug is tested. Furthermore, we demonstrate that our method is best suited for predicting DTIs in cases of drugs with no known targets or with few known targets. We also show the practical use of DASPfind by generating novel predictions for the Ion Channel dataset and validating them manually.
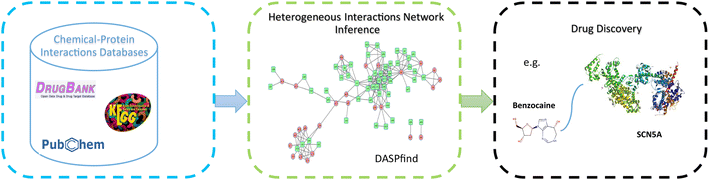
Conclusions: DASPfind is a computational method for finding reliable new interactions between drugs and proteins. We show over six different DTI datasets that DASPfind outperforms other state-of-the-art methods when the single top-ranked predictions are considered, or when a drug with no known targets or with few known targets is considered. We illustrate the usefulness and practicality of DASPfind by predicting novel DTIs for the Ion Channel dataset. The validated predictions suggest that DASPfind can be used as an efficient method to identify correct DTIs, thus reducing the cost of necessary experimental verifications in the process of drug discovery. DASPfind can be accessed online at: http://www.cbrc.kaust.edu.sa/daspfind.Graphical abstractThe conceptual workflow for predicting drug-target interactions using DASPfind.
Figures
References
-
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9(3):203–214. - PubMed
-
- CDER (2014) CDER’s 2013 novel new drugs. U.S. Food and Drug Administration. http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation/ucm38...
LinkOut - more resources
Full Text Sources
Other Literature Sources

