Automated screening for small organic ligands using DNA-encoded chemical libraries
- PMID: 26985574
- PMCID: PMC6126613
- DOI: 10.1038/nprot.2016.039
Automated screening for small organic ligands using DNA-encoded chemical libraries
Abstract
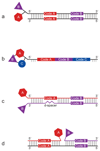
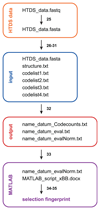
DNA-encoded chemical libraries (DECLs) are collections of organic compounds that are individually linked to different oligonucleotides, serving as amplifiable identification barcodes. As all compounds in the library can be identified by their DNA tags, they can be mixed and used in affinity-capture experiments on target proteins of interest. In this protocol, we describe the screening process that allows the identification of the few binding molecules within the multiplicity of library members. First, the automated affinity selection process physically isolates binding library members. Second, the DNA codes of the isolated binders are PCR-amplified and subjected to high-throughput DNA sequencing. Third, the obtained sequencing data are evaluated using a C++ program and the results are displayed using MATLAB software. The resulting selection fingerprints facilitate the discrimination of binding from nonbinding library members. The described procedures allow the identification of small organic ligands to biological targets from a DECL within 10 d.
Conflict of interest statement
D.N. is a co-founder and shareholder of Philochem AG (Otelfingen, Switzerland) and J.S. is a board member of Philochem AG.
Figures




References
-
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

