Characterization of Disease-Associated Mutations in Human Transmembrane Proteins
- PMID: 26986070
- PMCID: PMC4795776
- DOI: 10.1371/journal.pone.0151760
Characterization of Disease-Associated Mutations in Human Transmembrane Proteins
Abstract
Transmembrane protein coding genes are commonly associated with human diseases. We characterized disease causing mutations and natural polymorphisms in transmembrane proteins by mapping missense genetic variations from the UniProt database on the transmembrane protein topology listed in the Human Transmembrane Proteome database. We found characteristic differences in the spectrum of amino acid changes within transmembrane regions: in the case of disease associated mutations the non-polar to non-polar and non-polar to charged amino acid changes are equally frequent. In contrast, in the case of natural polymorphisms non-polar to charged amino acid changes are rare while non-polar to non-polar changes are common. The majority of disease associated mutations result in glycine to arginine and leucine to proline substitutions. Mutations to positively charged amino acids are more common in the center of the lipid bilayer, where they cause more severe structural and functional anomalies. Our analysis contributes to the better understanding of the effect of disease associated mutations in transmembrane proteins, which can help prioritize genetic variations in personal genomic investigations.
Conflict of interest statement
Figures
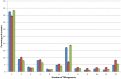
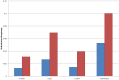

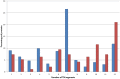
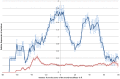
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

