In Vivo Molecular Dissection of the Effects of HIV-1 in Active Tuberculosis
- PMID: 26986567
- PMCID: PMC4795555
- DOI: 10.1371/journal.ppat.1005469
In Vivo Molecular Dissection of the Effects of HIV-1 in Active Tuberculosis
Abstract
Increased risk of tuberculosis (TB) associated with HIV-1 infection is primarily attributed to deficient T helper (Th)1 immune responses, but most people with active TB have robust Th1 responses, indicating that these are not sufficient to protect against disease. Recent findings suggest that favourable outcomes following Mycobacterium tuberculosis infection arise from finely balanced inflammatory and regulatory pathways, achieving pathogen control without immunopathology. We hypothesised that HIV-1 and antiretroviral therapy (ART) exert widespread changes to cell mediated immunity, which may compromise the optimal host protective response to TB and provide novel insights into the correlates of immune protection and pathogenesis. We sought to define these effects in patients with active TB by transcriptional profiling of tuberculin skin tests (TST) to make comprehensive molecular level assessments of in vivo human immune responses at the site of a standardised mycobacterial challenge. We showed that the TST transcriptome accurately reflects the molecular pathology at the site of human pulmonary TB, and used this approach to investigate immune dysregulation in HIV-1/TB co-infected patients with distinct clinical phenotypes associated with TST reactivity or anergy and unmasking TB immune reconstitution inflammatory syndrome (IRIS) after initiation of ART. HIV-1 infected patients with positive TSTs exhibited preserved Th1 responses but deficient immunoregulatory IL10-inducible responses. Those with clinically negative TSTs revealed profound anergy of innate as well as adaptive immune responses, except for preservation of type 1 interferon activity, implicated in impaired anti-mycobacterial immunity. Patients with unmasking TB IRIS showed recovery of Th1 immunity to normal levels, but exaggerated Th2-associated responses specifically. These mechanisms of immune dysregulation were localised to the tissue microenvironment and not evident in peripheral blood. TST molecular profiling categorised different mechanisms of immunological dysfunction in HIV-1 infection beyond the effects on CD4 T cells, each associated with increased risk of TB disease and amenable to host-directed therapies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
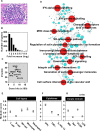




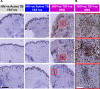
References
-
- WHO. WHO | Global tuberculosis report 2014 [Internet]. World Health Organisation; 2014. Available: http://www.who.int/tb/publications/global_report/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

