Evolution of Pentameric Ligand-Gated Ion Channels: Pro-Loop Receptors
- PMID: 26986966
- PMCID: PMC4795631
- DOI: 10.1371/journal.pone.0151934
Evolution of Pentameric Ligand-Gated Ion Channels: Pro-Loop Receptors
Abstract
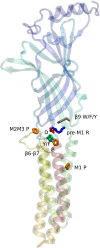
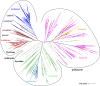
Pentameric ligand-gated ion channels (pLGICs) are ubiquitous neurotransmitter receptors in Bilateria, with a small number of known prokaryotic homologues. Here we describe a new inventory and phylogenetic analysis of pLGIC genes across all kingdoms of life. Our main finding is a set of pLGIC genes in unicellular eukaryotes, some of which are metazoan-like Cys-loop receptors, and others devoid of Cys-loop cysteines, like their prokaryotic relatives. A number of such "Cys-less" receptors also appears in invertebrate metazoans. Together, those findings draw a new distribution of pLGICs in eukaryotes. A broader distribution of prokaryotic channels also emerges, including a major new archaeal taxon, Thaumarchaeota. More generally, pLGICs now appear nearly ubiquitous in major taxonomic groups except multicellular plants and fungi. However, pLGICs are sparsely present in unicellular taxa, suggesting a high rate of gene loss and a non-essential character, contrasting with their essential role as synaptic receptors of the bilaterian nervous system. Multiple alignments of these highly divergent sequences reveal a small number of conserved residues clustered at the interface between the extracellular and transmembrane domains. Only the "Cys-loop" proline is absolutely conserved, suggesting the more fitting name "Pro loop" for that motif, and "Pro-loop receptors" for the superfamily. The infered molecular phylogeny shows a Cys-loop and a Cys-less clade in eukaryotes, both containing metazoans and unicellular members. This suggests new hypotheses on the evolutionary history of the superfamily, such as a possible origin of the Cys-loop cysteines in an ancient unicellular eukaryote. Deeper phylogenetic relationships remain uncertain, particularly around the split between bacteria, archaea, and eukaryotes.
Conflict of interest statement
Figures



References
-
- Ortells MO, Lunt GG. Evolutionary history of the ligand-gated ion-channel superfamily of receptors. Trends Neurosci. 1995. March;18(3):121–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

