Loss of miR-449a in ERG-associated prostate cancer promotes the invasive phenotype by inducing SIRT1
- PMID: 26988912
- PMCID: PMC5008401
- DOI: 10.18632/oncotarget.8061
Loss of miR-449a in ERG-associated prostate cancer promotes the invasive phenotype by inducing SIRT1
Abstract
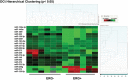
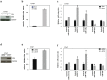
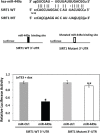
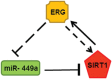
Epigenetic regulation by SIRT1, a multifaceted NAD+-dependent protein deacetylase, is one of the most common factors modulating cellular processes in a broad range of diseases, including prostate cancer (CaP). SIRT1 is over-expressed in CaP cells, however the associated mechanism is not well understood. To identify whether specific microRNAs might mediate this linkage, we have screened a miRNA library for differential expression in ERG-associated CaP tissues. Of 20 differentially and significantly expressed miRNAs that distinguish ERG-positive tumors from ERG-negative tumors, we find miR-449a is highly suppressed in ERG-positive tumors. We establish that SIRT1 is a direct target of miR-449a and is also induced by ERG in ERG-associated CaP. Our data suggest that attenuation of miR-449a promotes the invasive phenotype of the ERG-positive CaP in part by inducing the expression of SIRT1 in prostate cancer cells. Furthermore, we also find that suppression of SIRT1 results in a significant reduction in ERG expression in ERG-positive CaP cells, indicating a feed-back regulatory loop associated with ERG, miR-449a and SIRT1. We also report that ERG suppresses p53 acetylation perhaps through miR-449a-SIRT1 axis in CaP cells. Our findings provide new insight into the function of miRNAs in regulating ERG-associated CaP. Thus, miR-449a activation or SIRT1 suppression may represent new therapeutic opportunity for ERG-associated CaP.
Keywords: ERG; SIRT1; miR-449a; microRNA; prostate cancer.
Conflict of interest statement
All authors have no conflicts of interest in this study.
Figures





Similar articles
-
miR-449a targets HDAC-1 and induces growth arrest in prostate cancer.Oncogene. 2009 Apr 9;28(14):1714-24. doi: 10.1038/onc.2009.19. Epub 2009 Mar 2. Oncogene. 2009. PMID: 19252524
-
miR-449a enhances radiosensitivity through modulating pRb/E2F1 in prostate cancer cells.Tumour Biol. 2016 Apr;37(4):4831-40. doi: 10.1007/s13277-015-4336-8. Epub 2015 Oct 31. Tumour Biol. 2016. PMID: 26520443
-
miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells.Oncotarget. 2010 Sep;1(5):349-58. doi: 10.18632/oncotarget.167. Oncotarget. 2010. PMID: 20948989 Free PMC article.
-
miR-449a: a potential therapeutic agent for cancer.Anticancer Drugs. 2017 Nov;28(10):1067-1078. doi: 10.1097/CAD.0000000000000555. Anticancer Drugs. 2017. PMID: 29023247 Review.
-
Recent scenario of microRNA as diagnostic and prognostic biomarkers of prostate cancer.Urol Oncol. 2017 Mar;35(3):92-101. doi: 10.1016/j.urolonc.2016.10.019. Epub 2016 Nov 24. Urol Oncol. 2017. PMID: 27890424 Review.
Cited by
-
Modeling the role of microRNA-449a in the regulation of the G2/M cell cycle checkpoint in prostate LNCaP cells under ionizing radiation.PLoS One. 2018 Jul 19;13(7):e0200768. doi: 10.1371/journal.pone.0200768. eCollection 2018. PLoS One. 2018. PMID: 30024932 Free PMC article.
-
A Critical Review on microRNAs as Prognostic Biomarkers in Laryngeal Carcinoma.Int J Mol Sci. 2024 Dec 16;25(24):13468. doi: 10.3390/ijms252413468. Int J Mol Sci. 2024. PMID: 39769234 Free PMC article. Review.
-
miR-449a: A Promising Biomarker and Therapeutic Target in Cancer and Other Diseases.Cell Biochem Biophys. 2024 Sep;82(3):1629-1650. doi: 10.1007/s12013-024-01322-9. Epub 2024 May 29. Cell Biochem Biophys. 2024. PMID: 38809350 Review.
-
Molecular Sentinels: Unveiling the Role of Sirtuins in Prostate Cancer Progression.Int J Mol Sci. 2024 Dec 28;26(1):183. doi: 10.3390/ijms26010183. Int J Mol Sci. 2024. PMID: 39796040 Free PMC article. Review.
-
miR-449a Repression Leads to Enhanced NOTCH Signaling in TMPRSS2:ERG Fusion Positive Prostate Cancer Cells.Cancers (Basel). 2021 Feb 25;13(5):964. doi: 10.3390/cancers13050964. Cancers (Basel). 2021. PMID: 33669024 Free PMC article.
References
-
- Rosen P, Sesterhenn IA, Brassell SA, McLeod DG, Srivastava S, Dobi A. Clinical potential of the ERG oncoprotein in prostate cancer. Nature reviews Urology. 2012;9:131–137. - PubMed
-
- Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

