ATP1A3 Mutation in Adult Rapid-Onset Ataxia
- PMID: 26990090
- PMCID: PMC4798776
- DOI: 10.1371/journal.pone.0151429
ATP1A3 Mutation in Adult Rapid-Onset Ataxia
Abstract
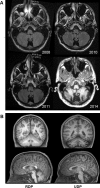
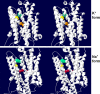
A 21-year old male presented with ataxia and dysarthria that had appeared over a period of months. Exome sequencing identified a de novo missense variant in ATP1A3, the gene encoding the α3 subunit of Na,K-ATPase. Several lines of evidence suggest that the variant is causative. ATP1A3 mutations can cause rapid-onset dystonia-parkinsonism (RDP) with a similar age and speed of onset, as well as severe diseases of infancy. The patient's ATP1A3 p.Gly316Ser mutation was validated in the laboratory by the impaired ability of the expressed protein to support the growth of cultured cells. In a crystal structure of Na,K-ATPase, the mutated amino acid was directly apposed to a different amino acid mutated in RDP. Clinical evaluation showed that the patient had many characteristics of RDP, however he had minimal fixed dystonia, a defining symptom of RDP. Successive magnetic resonance imaging (MRI) revealed progressive cerebellar atrophy, explaining the ataxia. The absence of dystonia in the presence of other RDP symptoms corroborates other evidence that the cerebellum contributes importantly to dystonia pathophysiology. We discuss the possibility that a second de novo variant, in ubiquilin 4 (UBQLN4), a ubiquitin pathway component, contributed to the cerebellar neurodegenerative phenotype and differentiated the disease from other manifestations of ATP1A3 mutations. We also show that a homozygous variant in GPRIN1 (G protein-regulated inducer of neurite outgrowth 1) deletes a motif with multiple copies and is unlikely to be causative.
Conflict of interest statement
Figures

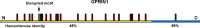
References
-
- Brashear A, Sweadner KJ, Cook JF, Swoboda KJ, Ozelius LJ (2014) ATP1A3-related neurologic disorders In: Pagon RA, Adam MP, Ardinger HH, et al. , editors. GeneReviews. NCBI Bookshelf.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

