Entamoeba Clone-Recognition Experiments: Morphometrics, Aggregative Behavior, and Cell-Signaling Characterization
- PMID: 26990199
- PMCID: PMC4856553
- DOI: 10.1111/jeu.12313
Entamoeba Clone-Recognition Experiments: Morphometrics, Aggregative Behavior, and Cell-Signaling Characterization
Abstract
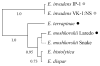
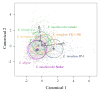
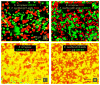
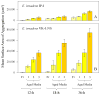
Studies on clone- and kin-discrimination in protists have proliferated during the past decade. We report clone-recognition experiments in seven Entamoeba lineages (E. invadens IP-1, E. invadens VK-1:NS, E. terrapinae, E. moshkovskii Laredo, E. moshkovskii Snake, E. histolytica HM-1:IMSS and E. dispar). First, we characterized morphometrically each clone (length, width, and cell-surface area) and documented how they differed statistically from one another (as per single-variable or canonical-discriminant analyses). Second, we demonstrated that amebas themselves could discriminate self (clone) from different (themselves vs. other clones). In mix-cell-line cultures between closely-related (E. invadens IP-1 vs. E. invadens VK-1:NS) or distant-phylogenetic clones (E. terrapinae vs. E. moshkovskii Laredo), amebas consistently aggregated with same-clone members. Third, we identified six putative cell-signals secreted by the amebas (RasGap/Ankyrin, coronin-WD40, actin, protein kinases, heat shock 70, and ubiquitin) and which known functions in Entamoeba spp. included: cell proliferation, cell adhesion, cell movement, and stress-induced encystation. To our knowledge, this is the first multi-clone characterization of Entamoeba spp. morphometrics, aggregative behavior, and cell-signaling secretion in the context of clone-recognition. Protists allow us to study cell-cell recognition from ecological and evolutionary perspectives. Modern protistan lineages can be central to studies about the origins and evolution of multicellularity.
Keywords: Active cell death; Entamoeba proliferation activating factors; cell migration; chemical cues; inclusive fitness; social ameba.
© 2016 The Author(s) Journal of Eukaryotic Microbiology © 2016 International Society of Protistologists.
Figures


References
-
- Biedrzycki ML, Bais HP. Kin recognition in plants: a mysterious behaviour unsolved. J Exp Bot. 2010;61:4123–4128. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

