Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis)
- PMID: 26992010
- PMCID: PMC4877267
- DOI: 10.1111/mec.13605
Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis)
Abstract
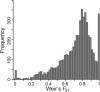
The evolutionary mechanisms generating the tremendous biodiversity of islands have long fascinated evolutionary biologists. Genetic drift and divergent selection are predicted to be strong on islands and both could drive population divergence and speciation. Alternatively, strong genetic drift may preclude adaptation. We conducted a genomic analysis to test the roles of genetic drift and divergent selection in causing genetic differentiation among populations of the island fox (Urocyon littoralis). This species consists of six subspecies, each of which occupies a different California Channel Island. Analysis of 5293 SNP loci generated using Restriction-site Associated DNA (RAD) sequencing found support for genetic drift as the dominant evolutionary mechanism driving population divergence among island fox populations. In particular, populations had exceptionally low genetic variation, small Ne (range = 2.1-89.7; median = 19.4), and significant genetic signatures of bottlenecks. Moreover, islands with the lowest genetic variation (and, by inference, the strongest historical genetic drift) were most genetically differentiated from mainland grey foxes, and vice versa, indicating genetic drift drives genome-wide divergence. Nonetheless, outlier tests identified 3.6-6.6% of loci as high FST outliers, suggesting that despite strong genetic drift, divergent selection contributes to population divergence. Patterns of similarity among populations based on high FST outliers mirrored patterns based on morphology, providing additional evidence that outliers reflect adaptive divergence. Extremely low genetic variation and small Ne in some island fox populations, particularly on San Nicolas Island, suggest that they may be vulnerable to fixation of deleterious alleles, decreased fitness and reduced adaptive potential.
Keywords: conservation genomics; divergent selection; effective population size; genetic drift; population divergence.
© 2016 John Wiley & Sons Ltd.
Figures




References
-
- Balkenhol N, Waits LP, Dezzani RJ. Statistical approaches in landscape genetics: an evaluation of methods for linking landscape and genetic data. Ecography. 2009;32:818–830.
-
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13:969–980. - PubMed
-
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society B-Biological Sciences. 1996;263:1619–1626.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

