Using Supervised Machine Learning to Classify Real Alerts and Artifact in Online Multisignal Vital Sign Monitoring Data
- PMID: 26992068
- PMCID: PMC4911247
- DOI: 10.1097/CCM.0000000000001660
Using Supervised Machine Learning to Classify Real Alerts and Artifact in Online Multisignal Vital Sign Monitoring Data
Abstract
Objective: The use of machine-learning algorithms to classify alerts as real or artifacts in online noninvasive vital sign data streams to reduce alarm fatigue and missed true instability.
Design: Observational cohort study.
Setting: Twenty-four-bed trauma step-down unit.
Patients: Two thousand one hundred fifty-three patients.
Intervention: Noninvasive vital sign monitoring data (heart rate, respiratory rate, peripheral oximetry) recorded on all admissions at 1/20 Hz, and noninvasive blood pressure less frequently, and partitioned data into training/validation (294 admissions; 22,980 monitoring hours) and test sets (2,057 admissions; 156,177 monitoring hours). Alerts were vital sign deviations beyond stability thresholds. A four-member expert committee annotated a subset of alerts (576 in training/validation set, 397 in test set) as real or artifact selected by active learning, upon which we trained machine-learning algorithms. The best model was evaluated on test set alerts to enact online alert classification over time.
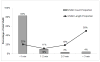
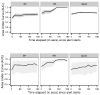
Measurements and main results: The Random Forest model discriminated between real and artifact as the alerts evolved online in the test set with area under the curve performance of 0.79 (95% CI, 0.67-0.93) for peripheral oximetry at the instant the vital sign first crossed threshold and increased to 0.87 (95% CI, 0.71-0.95) at 3 minutes into the alerting period. Blood pressure area under the curve started at 0.77 (95% CI, 0.64-0.95) and increased to 0.87 (95% CI, 0.71-0.98), whereas respiratory rate area under the curve started at 0.85 (95% CI, 0.77-0.95) and increased to 0.97 (95% CI, 0.94-1.00). Heart rate alerts were too few for model development.
Conclusions: Machine-learning models can discern clinically relevant peripheral oximetry, blood pressure, and respiratory rate alerts from artifacts in an online monitoring dataset (area under the curve > 0.87).
Figures


Comment in
-
Computers in White Coats: How to Devise Useful Clinical Decision Support Software.Crit Care Med. 2016 Jul;44(7):1449-50. doi: 10.1097/CCM.0000000000001781. Crit Care Med. 2016. PMID: 27309173 No abstract available.
References
-
- Otero A, Félix P, Barro S, Palacios F. Addressing the flaws of current critical alarms: a fuzzy constraint satisfaction approach. Artif Intell Med. 2009;47:219–238. - PubMed
-
- Takla G, Petre JH, Doyle DJ, Horibe M, Gopakumaran B. The problem of artifacts in patient monitor data during surgery: a clinical and methodological review. Anest Analg. 2006;103:1196–1204. - PubMed
-
- Smith M. Rx for ECG monitoring artifact. Crit Care Nurse. 1984;4:64. - PubMed
-
- Boumbarov O, Velchev Y, Sokolov S. ECG personal identification in subspaces using radial basis neural networks. Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications. 2009 IDAACS 2009 IEEE International Workshop on IEEE; 2009. pp. 446–451.
-
- Paul JS, Reddy MR, Kumar VJ. A transform domain SVD filter for suppression of muscle noise artifacts in exercise ECG’s. IEEE Trans Biomed Eng. 2000;47:654–663. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

