Downregulation of the stress-induced ligand ULBP1 following SV40 infection confers viral evasion from NK cell cytotoxicity
- PMID: 26992229
- PMCID: PMC4941247
- DOI: 10.18632/oncotarget.8085
Downregulation of the stress-induced ligand ULBP1 following SV40 infection confers viral evasion from NK cell cytotoxicity
Abstract
Polyomaviruses are a diverse family of viruses which are prevalent in the human population. However, the interactions of these viruses with the immune system are not well characterized. We have previously shown that two human polyomaviruses, JC and BK, use an identical microRNA to evade immune attack by Natural Killer (NK) cells. We showed that this viral microRNA suppresses ULBP3 expression, a stress induced ligand for the killer receptor NKG2D. Here we show that Simian Virus 40 (SV40) also evades NK cell attack through the down regulation of another stress-induced ligand of NKG2D, ULBP1. These findings indicate that NK cells play an essential role in fighting polyomavirus infections and further emphasize the importance of various members of the ULBP family in controlling polyomavirus infection.
Keywords: Immune response; Immunity; Immunology and Microbiology Section; NK cells; NKG2D; SV40; ULBP1; immune-evasion.
Conflict of interest statement
The authors have no conflict of interest.
Figures

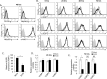



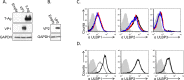

References
-
- Arnon TI, Markel G, Mandelboim O. Tumor and viral recognition by natural killer cells receptors. Seminars in cancer biology. 2006;16:348–358. - PubMed
-
- Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunology and cell biology. 2011;89:216–224. - PubMed
-
- Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends in immunology. 2005;26:221–226. - PubMed
-
- Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

