RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence
- PMID: 26992231
- PMCID: PMC4991368
- DOI: 10.18632/oncotarget.8087
RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence
Abstract
The tRNA methyltransferase NSUN2 delays replicative senescence by regulating the translation of CDK1 and CDKN1B mRNAs. However, whether NSUN2 influences premature cellular senescence remains untested. Here we show that NSUN2 methylates SHC mRNA in vitro and in cells, thereby enhancing the translation of the three SHC proteins, p66SHC, p52SHC, and p46SHC. Our results further show that the elevation of SHC expression by NSUN2-mediated mRNA methylation increased the levels of ROS, activated p38MAPK, thereby accelerating oxidative stress- and high-glucose-induced senescence of human vascular endothelial cells (HUVEC). Our findings highlight the critical impact of NSUN2-mediated mRNA methylation in promoting premature senescence.
Keywords: Gerotarget; HUVEC; NSUN2; SHC mRNA methylation; premature senescence; translational regulation.
Conflict of interest statement
We declare no conflicts of interest.
Figures

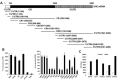
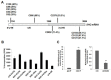
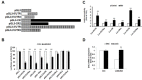


References
-
- Luzi L, Confalonieri S, Di Fiore PP, Pelicci PG. Evolution of SHC functions from nematode to human. Curr Opin Genet Dev. 2000;10:668–674. - PubMed
-
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66SHC adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. - PubMed
-
- Napoli C, Martin-Padura I, de Nigris F, Giorgio M, Mansueto G, Somma P, Condorelli M, Sica G, De Rosa G, Pelicci P. Deletion of the p66SHC longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc Natl Acad Sci U S A. 2003;100:2112–2116. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

