Tumour-associated and non-tumour-associated microbiota in colorectal cancer
- PMID: 26992426
- PMCID: PMC5529966
- DOI: 10.1136/gutjnl-2015-309595
Tumour-associated and non-tumour-associated microbiota in colorectal cancer
Abstract
Objective: A signature that unifies the colorectal cancer (CRC) microbiota across multiple studies has not been identified. In addition to methodological variance, heterogeneity may be caused by both microbial and host response differences, which was addressed in this study.
Design: We prospectively studied the colonic microbiota and the expression of specific host response genes using faecal and mucosal samples ('ON' and 'OFF' the tumour, proximal and distal) from 59 patients undergoing surgery for CRC, 21 individuals with polyps and 56 healthy controls. Microbiota composition was determined by 16S rRNA amplicon sequencing; expression of host genes involved in CRC progression and immune response was quantified by real-time quantitative PCR.
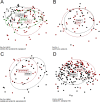
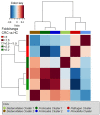
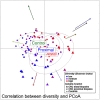
Results: The microbiota of patients with CRC differed from that of controls, but alterations were not restricted to the cancerous tissue. Differences between distal and proximal cancers were detected and faecal microbiota only partially reflected mucosal microbiota in CRC. Patients with CRC can be stratified based on higher level structures of mucosal-associated bacterial co-abundance groups (CAGs) that resemble the previously formulated concept of enterotypes. Of these, Bacteroidetes Cluster 1 and Firmicutes Cluster 1 were in decreased abundance in CRC mucosa, whereas Bacteroidetes Cluster 2, Firmicutes Cluster 2, Pathogen Cluster and Prevotella Cluster showed increased abundance in CRC mucosa. CRC-associated CAGs were differentially correlated with the expression of host immunoinflammatory response genes.
Conclusions: CRC-associated microbiota profiles differ from those in healthy subjects and are linked with distinct mucosal gene-expression profiles. Compositional alterations in the microbiota are not restricted to cancerous tissue and differ between distal and proximal cancers.
Keywords: COLORECTAL CANCER; GENE EXPRESSION; INTESTINAL MICROBIOLOGY.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/.
Conflict of interest statement
Figures






Comment in
-
Tumour-associated and non-tumour-associated microbiota in colorectal cancer.Gut. 2018 Feb;67(2):395. doi: 10.1136/gutjnl-2017-314219. Epub 2017 May 4. Gut. 2018. PMID: 28473629 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
