Confirmation of a non-synonymous SNP in PNPLA8 as a candidate causal mutation for Weaver syndrome in Brown Swiss cattle
- PMID: 26992691
- PMCID: PMC4797220
- DOI: 10.1186/s12711-016-0201-5
Confirmation of a non-synonymous SNP in PNPLA8 as a candidate causal mutation for Weaver syndrome in Brown Swiss cattle
Abstract
Background: Bovine progressive degenerative myeloencephalopathy (Weaver syndrome) is a neurodegenerative disorder in Brown Swiss cattle that is characterized by progressive hind leg weakness and ataxia, while sensorium and spinal reflexes remain unaffected. Although the causal mutation has not been identified yet, an indirect genetic test based on six microsatellite markers and consequent exclusion of Weaver carriers from breeding have led to the complete absence of new cases for over two decades. Evaluation of disease status by imputation of 41 diagnostic single nucleotide polymorphisms (SNPs) and a common haplotype published in 2013 identified several suspected carriers in the current breeding population, which suggests a higher frequency of the Weaver allele than anticipated. In order to prevent the reemergence of the disease, this study aimed at mapping the gene that underlies Weaver syndrome and thus at providing the basis for direct genetic testing and monitoring of today's Braunvieh/Brown Swiss herds.
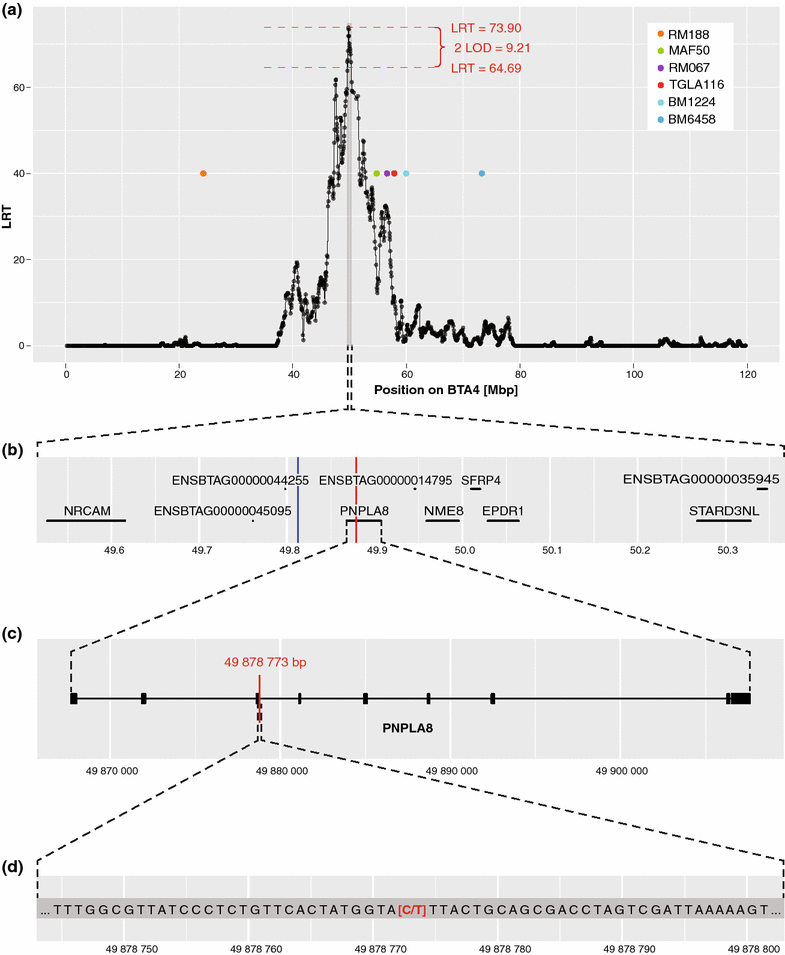
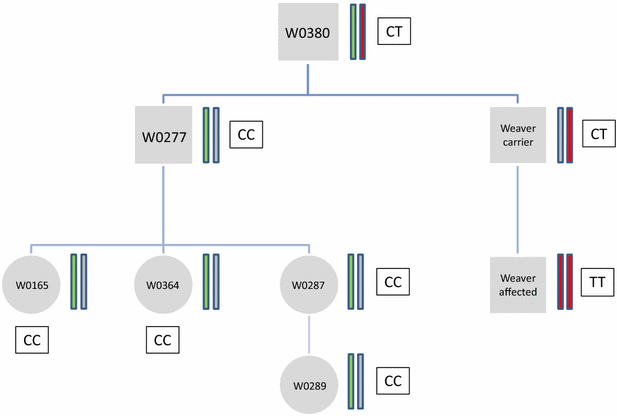
Results: Combined linkage/linkage disequilibrium mapping on Bos taurus chromosome (BTA) 4 based on Illumina Bovine SNP50 genotypes of 43 Weaver-affected, 31 Weaver carrier and 86 Weaver-free animals resulted in a maximum likelihood ratio test statistic value at position 49,812,384 bp. The confidence interval (0.853 Mb) determined by the 2-LOD drop-off method was contained within a 1.72-Mb segment of extended homozygosity. Exploitation of whole-genome sequence data from two official Weaver carriers and 1145 other bulls that were sequenced in Run4 of the 1000 bull genomes project showed that only a non-synonymous SNP (rs800397662) within the PNPLA8 gene at position 49,878,773 bp was concordant with the Weaver carrier status. Targeted SNP genotyping confirmed this SNP as a candidate causal mutation for Weaver syndrome. Genotyping for the candidate causal mutation in a random sample of 2334 current Braunvieh animals suggested a frequency of the Weaver allele of 0.26 %.
Conclusions: Through combined use of exhaustive sequencing data and SNP genotyping results, we were able to provide evidence that supports the non-synonymous mutation at position 49,878,773 bp as the most likely causal mutation for Weaver syndrome. Further studies are needed to uncover the exact mechanisms that underlie this syndrome.
Figures

References
-
- Leipold HW, Blaugh B, Huston K, Edgerly CG, Hibbs CM. Weaver syndrome in Brown Swiss cattle: clinical signs and pathology. Vet Med Small Anim Clin. 1973;68:645–647. - PubMed
-
- Drögemüller C, Tetens J, Sigurdsson S, Gentile A, Testoni S, Lindblad-Toh K, et al. Identification of the bovine Arachnomelia mutation by massively parallel sequencing implicates sulfite oxidase (SUOX) in bone development. PLoS Genet. 2010;6:e1001079. doi: 10.1371/journal.pgen.1001079. - DOI - PMC - PubMed
-
- Trela T. Untersuchungen über Klinik und Verlauf der Bovinen progressiv-degenerativen Myeloenzephlopathie (“Weaver-Syndrom”) bei Brown-Swiss × Braunviehrindern. Inaugural-Dissertation, Ludwig-Maximilians-Universität. 1991.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

