Systems biology of lens development: A paradigm for disease gene discovery in the eye
- PMID: 26992779
- PMCID: PMC5026553
- DOI: 10.1016/j.exer.2016.03.010
Systems biology of lens development: A paradigm for disease gene discovery in the eye
Abstract
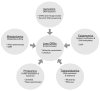
Over the past several decades, the biology of the developing lens has been investigated using molecular genetics-based approaches in various vertebrate model systems. These efforts, involving target gene knockouts or knockdowns, have led to major advances in our understanding of lens morphogenesis and the pathological basis of cataracts, as well as of other lens related eye defects. In particular, we now have a functional understanding of regulators such as Pax6, Six3, Sox2, Oct1 (Pou2f1), Meis1, Pnox1, Zeb2 (Sip1), Mab21l1, Foxe3, Tfap2a (Ap2-alpha), Pitx3, Sox11, Prox1, Sox1, c-Maf, Mafg, Mafk, Hsf4, Fgfrs, Bmp7, and Tdrd7 in this tissue. However, whether these individual regulators interact or their targets overlap, and the significance of such interactions during lens morphogenesis, is not well defined. The arrival of high-throughput approaches for gene expression profiling (microarrays, RNA-sequencing (RNA-seq), etc.), which can be coupled with chromatin immunoprecipitation (ChIP) or RNA immunoprecipitation (RIP) assays, along with improved computational resources and publically available datasets (e.g. those containing comprehensive protein-protein, protein-DNA information), presents new opportunities to advance our understanding of the lens tissue on a global systems level. Such systems-level knowledge will lead to the derivation of the underlying lens gene regulatory network (GRN), defined as a circuit map of the regulator-target interactions functional in lens development, which can be applied to expedite cataract gene discovery. In this review, we cover the various systems-level approaches such as microarrays, RNA-seq, and ChIP that are already being applied to lens studies and discuss strategies for assembling and interpreting these vast amounts of high-throughput information for effective dispersion to the scientific community. In particular, we discuss strategies for effective interpretation of this new information in the context of the rich knowledge obtained through the application of traditional single-gene focused experiments on the lens. Finally, we discuss our vision for integrating these diverse high-throughput datasets in a single web-based user-friendly tool iSyTE (integrated Systems Tool for Eye gene discovery) - a resource that is already proving effective in the identification and characterization of genes linked to lens development and cataract. We anticipate that application of a similar approach to other ocular tissues such as the retina and the cornea, and even other organ systems, will significantly impact disease gene discovery.
Keywords: Bioinformatics; Cataract; Gene regulatory networks; Lens; RNA binding proteins; Transcription factors; iSyTE.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures


References
-
- Agrawal SA, Anand D, Siddam AD, Kakrana A, Dash S, Scheiblin DA, Dang CA, Terrell AM, Waters SM, Singh A, Motohashi H, Yamamoto M, Lachke SA. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet. 2015;134:717–735. doi: 10.1007/s00439-015-1554-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

