The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations
- PMID: 26992833
- PMCID: PMC5126213
- DOI: 10.15252/emmm.201506055
The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations
Abstract
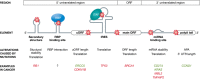
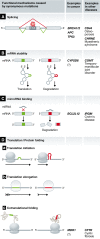
Cancer is a disease of the genome caused by oncogene activation and tumor suppressor gene inhibition. Deep sequencing studies including large consortia such as TCGA and ICGC identified numerous tumor-specific mutations not only in protein-coding sequences but also in non-coding sequences. Although 98% of the genome is not translated into proteins, most studies have neglected the information hidden in this "dark matter" of the genome. Malignancy-driving mutations can occur in all genetic elements outside the coding region, namely in enhancer, silencer, insulator, and promoter as well as in 5'-UTR and 3'-UTR Intron or splice site mutations can alter the splicing pattern. Moreover, cancer genomes contain mutations within non-coding RNA, such as microRNA, lncRNA, and lincRNA A synonymous mutation changes the coding region in the DNA and RNA but not the protein sequence. Importantly, oncogenes such as TERT or miR-21 as well as tumor suppressor genes such as TP53/p53, APC, BRCA1, or RB1 can be affected by these alterations. In summary, coding-independent mutations can affect gene regulation from transcription, splicing, mRNA stability to translation, and hence, this largely neglected area needs functional studies to elucidate the mechanisms underlying tumorigenesis. This review will focus on the important role and novel mechanisms of these non-coding or allegedly silent mutations in tumorigenesis.
Keywords: alternative polyadenylation; enhancer; mutation; non‐coding RNA; synonymous mutation.
© 2016 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

References
-
- Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB (2010) Annotating non‐coding regions of the genome. Nat Rev Genet 11: 559–571 - PubMed
-
- Anczukow O, Buisson M, Leone M, Coutanson C, Lasset C, Calender A, Sinilnikova OM, Mazoyer S (2012) BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin Cancer Res 18: 4903–4909 - PubMed
-
- Anczuków O, Buisson M, Salles M‐J, Triboulet S, Longy M, Lidereau R, Sinilnikova OM, Mazoyer S (2008) Unclassified variants identified in BRCA1 exon 11: consequences on splicing. Genes Chromosom Cancer 47: 418–426 - PubMed
-
- Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, Struewing JP, Stoppa‐Lyonnet D, Barjhoux L, Hughes DJ et al (2007) RAD51 135G–>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet 81: 1186–1200 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

