Kainic Acid Induces mTORC1-Dependent Expression of Elmo1 in Hippocampal Neurons
- PMID: 26993296
- PMCID: PMC5390005
- DOI: 10.1007/s12035-016-9821-6
Kainic Acid Induces mTORC1-Dependent Expression of Elmo1 in Hippocampal Neurons
Abstract
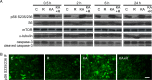
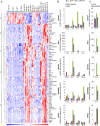
Epileptogenesis is a process triggered by initial environmental or genetic factors that result in epilepsy and may continue during disease progression. Important parts of this process include changes in transcriptome and the pathological rewiring of neuronal circuits that involves changes in neuronal morphology. Mammalian/mechanistic target of rapamycin (mTOR) is upregulated by proconvulsive drugs, e.g., kainic acid, and is needed for progression of epileptogenesis, but molecular aspects of its contribution are not fully understood. Since mTOR can modulate transcription, we tested if rapamycin, an mTOR complex 1 inhibitor, affects kainic acid-evoked transcriptome changes. Using microarray technology, we showed that rapamycin inhibits the kainic acid-induced expression of multiple functionally heterogeneous genes. We further focused on engulfment and cell motility 1 (Elmo1), which is a modulator of actin dynamics and therefore could contribute to pathological rewiring of neuronal circuits during epileptogenesis. We showed that prolonged overexpression of Elmo1 in cultured hippocampal neurons increased axonal growth, decreased dendritic spine density, and affected their shape. In conclusion, data presented herein show that increased mTORC1 activity in response to kainic acid has no global effect on gene expression. Instead, our findings suggest that mTORC1 inhibition may affect development of epilepsy, by modulating expression of specific subset of genes, including Elmo1, and point to a potential role for Elmo1 in morphological changes that accompany epileptogenesis.
Keywords: Elmo1; Epilepsy; Gene expression; Rapamycin; mTORC1.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

