Inactivation of RASA1 promotes melanoma tumorigenesis via R-Ras activation
- PMID: 26993606
- PMCID: PMC5029671
- DOI: 10.18632/oncotarget.8127
Inactivation of RASA1 promotes melanoma tumorigenesis via R-Ras activation
Abstract
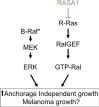
Inactivation of Ras GTPase activating proteins (RasGAPs) can activate Ras, increasing the risk for tumor development. Utilizing a melanoma whole genome sequencing (WGS) data from 13 patients, we identified two novel, clustered somatic missense mutations (Y472H and L481F) in RASA1 (RAS p21 protein activator 1, also called p120RasGAP). We have shown that wild type RASA1, but not identified mutants, suppresses soft agar colony formation and tumor growth of BRAF mutated melanoma cell lines via its RasGAP activity toward R-Ras (related RAS viral (r-ras) oncogene homolog) isoform. Moreover, R-Ras increased and RASA1 suppressed Ral-A activation among Ras downstream effectors. In addition to mutations, loss of RASA1 expression was frequently observed in metastatic melanoma samples on melanoma tissue microarray (TMA) and a low level of RASA1 mRNA expression was associated with decreased overall survival in melanoma patients with BRAF mutations. Thus, these data support that RASA1 is inactivated by mutation or by suppressed expression in melanoma and that RASA1 plays a tumor suppressive role by inhibiting R-Ras, a previously less appreciated member of the Ras small GTPases.
Keywords: R-Ras; RASA1; RasGAP; melanoma; whole genome sequencing.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

