Characterisation of the epidemic strain of H3N8 equine influenza virus responsible for outbreaks in South America in 2012
- PMID: 26993620
- PMCID: PMC4799594
- DOI: 10.1186/s12985-016-0503-9
Characterisation of the epidemic strain of H3N8 equine influenza virus responsible for outbreaks in South America in 2012
Abstract
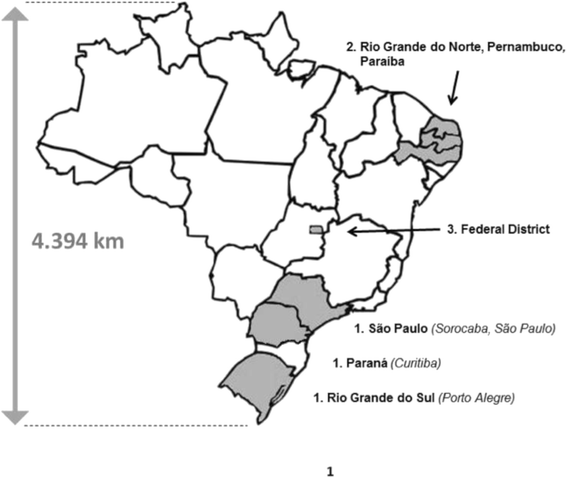
Background: An extensive outbreak of equine influenza occurred across multiple countries in South America during 2012. The epidemic was first reported in Chile then spread to Brazil, Uruguay and Argentina, where both vaccinated and unvaccinated animals were affected. In Brazil, infections were widespread within 3months of the first reported cases. Affected horses included animals vaccinated with outdated vaccine antigens, but also with the OIE-recommended Florida clade 1 strain South Africa/4/03.
Methods: Equine influenza virus strains from infected horses were isolated in eggs, then a representative strain was subjected to full genome sequencing using segment-specific primers with M13 tags. Phylogenetic analyses of nucleotide sequences were completed using PhyML. Amino acid sequences of haemagglutinin and neuraminidase were compared against those of vaccine strains and recent isolates from America and Uruguay, substitutions were mapped onto 3D protein structures using PyMol. Antigenic analyses were completed by haemagglutination-inhibition assay using post-infection ferret sera.
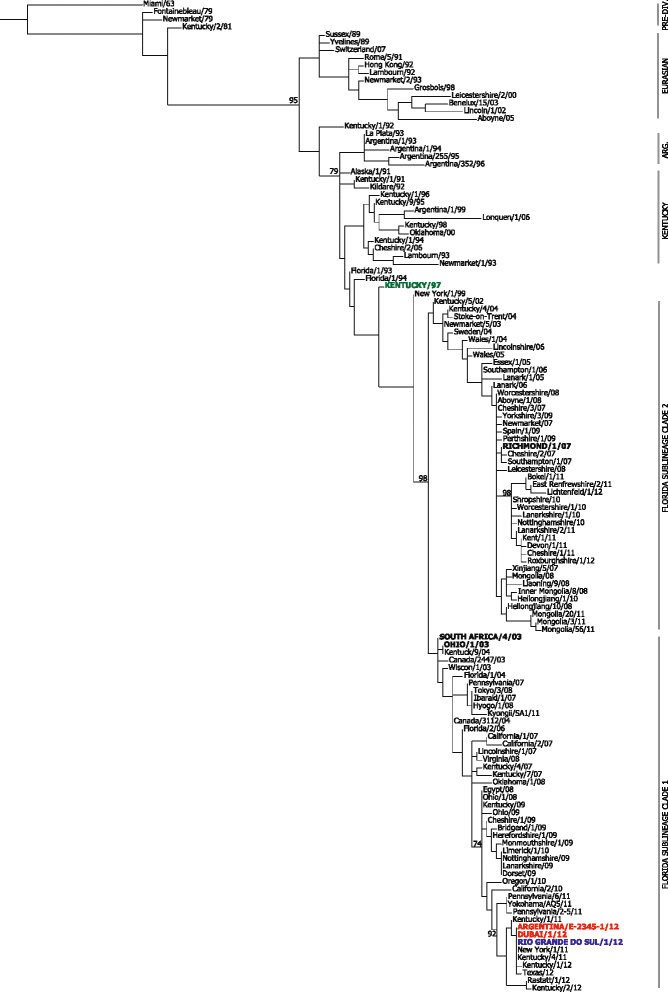
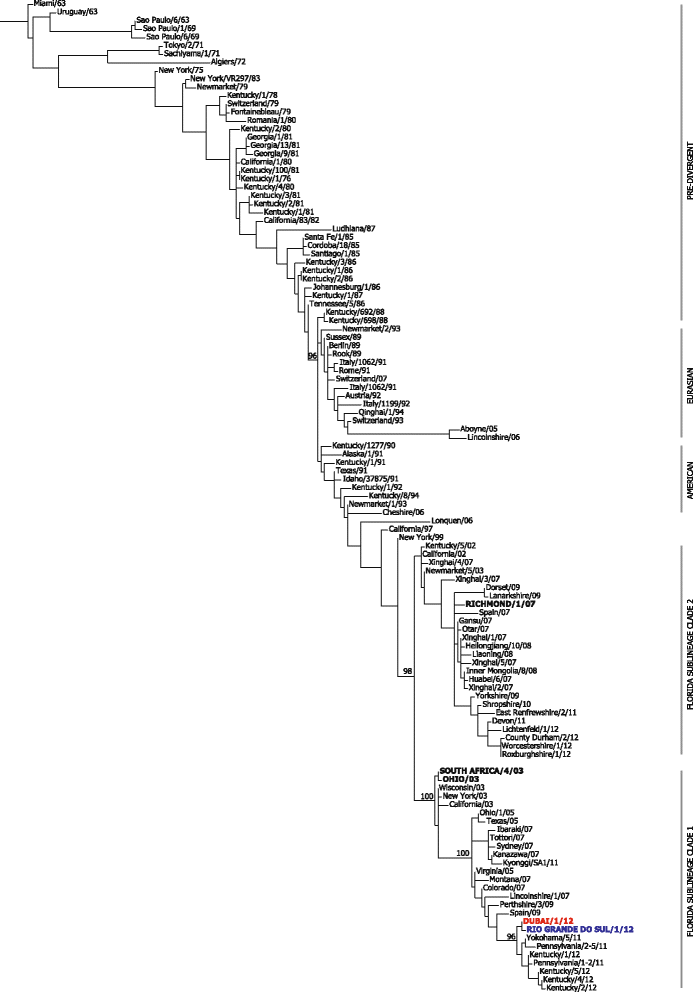
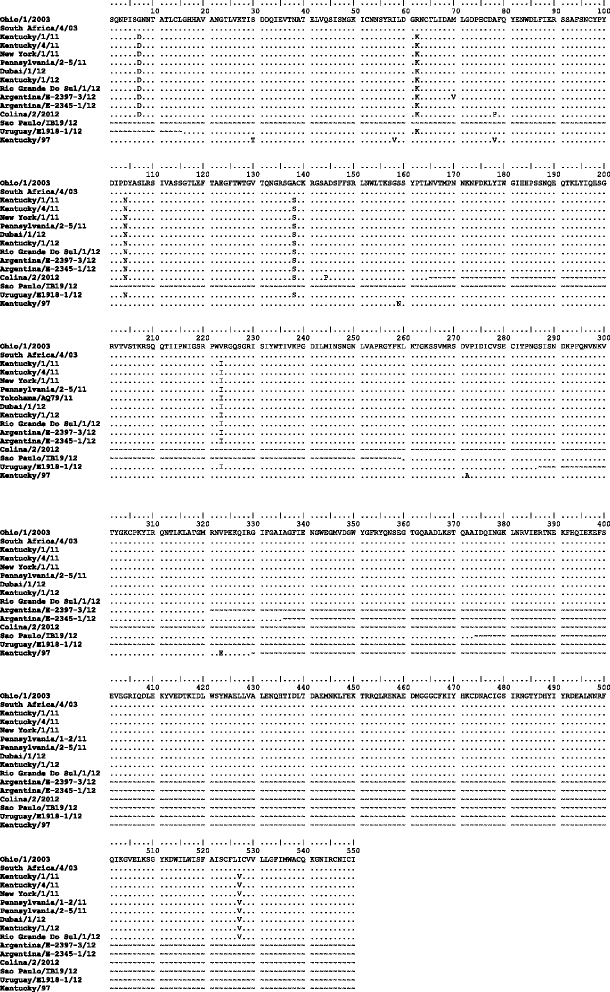
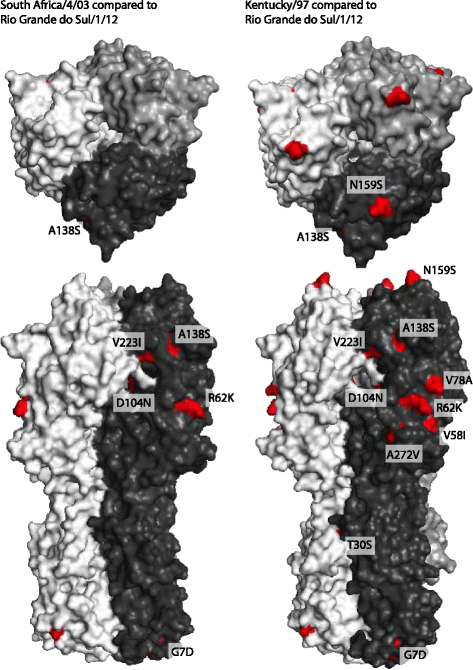
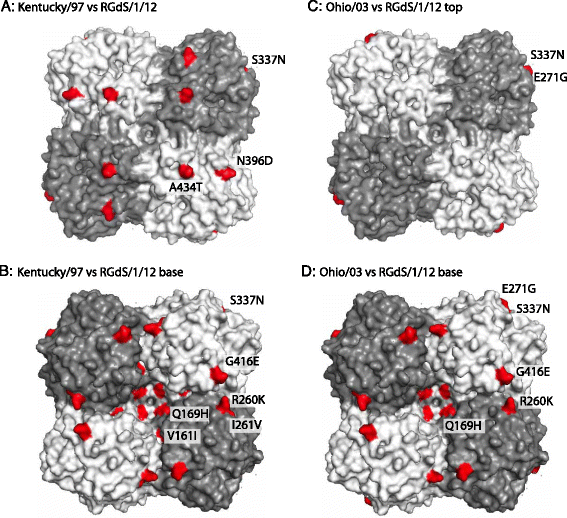
Results: Nucleotide sequences of the haemaglutinin (HA) and neuraminidase (NA) genes of Brazilian isolate A/equine/Rio Grande do Sul/2012 were very similar to those of viruses belonging to Florida clade 1 and clustered with contemporary isolates from the USA. Comparison of their amino acid sequences against the OIE-recommended Florida clade 1 vaccine strain A/equine/South Africa/4/03 revealed five amino acid substitutions in HA and seven in NA. Changes in HA included one within antigenic site A and one within the 220-loop of the sialic acid receptor binding site. However, antigenic analysis by haemagglutination inhibition (HI) assay with ferret antisera raised against representatives of European, Kentucky and Florida sublineages failed to indicate any obvious differences in antigenicity.
Conclusions: An extensive outbreak of equine influenza in South America during 2012 was caused by a virus belonging to Florida clade 1, closely related to strains circulating in the USA in 2011. Despite reports of vaccine breakdown with products containing the recommended strain South Africa/03, no evidence was found of significant antigenic drift. Other factors may have contributed to the rapid spread of this virus, including poor control of horse movement.
Keywords: Brazil; Epidemic; Equine influenza; Florida clade 1; Haemagglutinin; Neuraminidase.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

