PIM1 destabilization activates a p53-dependent response to ribosomal stress in cancer cells
- PMID: 26993775
- PMCID: PMC5029667
- DOI: 10.18632/oncotarget.8070
PIM1 destabilization activates a p53-dependent response to ribosomal stress in cancer cells
Abstract
Defects in ribosome biogenesis triggers a stress response (ribosomal stress) that can lead to growth arrest and apoptosis. Signaling pathways activated by ribosomal stress are specifically involved in the pathological mechanism of a group of disorders defined as ribosomopathies. However, more generally, the quality control of ribosome synthesis is part of the regulatory circuits that control cell metabolism. A number of studies identified tumor suppressor p53 as a central player in ribosomal stress. We have previously reported that the kinase PIM1 plays a role as a sensor for ribosome deficiency. In this report we address the relationship between PIM1 and p53 in cancer cell lines after depletion of a ribosomal protein. We identified a novel signaling pathway that includes the kinase AKT and the ubiquitin ligase MDM2. In fact, our results indicate that the lower level of PIM1, induced by ribosomal stress, causes inactivation of AKT, inhibition of MDM2 and a consequent p53 stabilization. Therefore, we propose that activation of p53 in response to ribosomal stress, is dependent on the pathway PIM1-AKT-MDM2. In addition, we report evidence that PIM1 level may be relevant to assess the sensitivity of cancer cells to chemotherapeutic drugs that induce ribosomal stress.
Keywords: AKT; MDM2; PIM1; p53; ribosomal stress.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





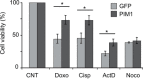
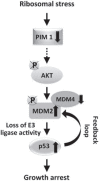
References
-
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. - PubMed
-
- Dianzani I, Loreni F. Diamond-Blackfan anemia: a ribosomal puzzle. Haematologica. 2008;93:1601–1604. - PubMed
-
- Loreni F, Mancino M, Biffo S. Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene. 2014;33:2145–2156. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

