Silica-Induced Protein (Sip) in Thermophilic Bacterium Thermus thermophilus Responds to Low Iron Availability
- PMID: 26994077
- PMCID: PMC4959228
- DOI: 10.1128/AEM.04027-15
Silica-Induced Protein (Sip) in Thermophilic Bacterium Thermus thermophilus Responds to Low Iron Availability
Abstract
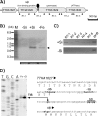
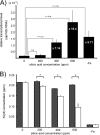
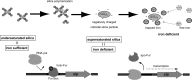
Thermus thermophilus HB8 expresses silica-induced protein (Sip) when cultured in medium containing supersaturated silicic acids. Using genomic information, Sip was identified as a Fe(3+)-binding ABC transporter. Detection of a 1-kb hybridized band in Northern analysis revealed that sip transcription is monocistronic and that sip has its own terminator and promoter. The sequence of the sip promoter showed homology with that of the σ(A)-dependent promoter, which is known as a housekeeping promoter in HB8. Considering that sip is transcribed when supersaturated silicic acids are added, the existence of a repressor is presumed. DNA microarray analysis suggested that supersaturated silicic acids and iron deficiency affect Thermus cells similarly, and enhanced sip transcription was detected under both conditions. This suggested that sip transcription was initiated by iron deficiency and that the ferric uptake regulator (Fur) controlled the transcription. Three Fur gene homologues (TTHA0255, TTHA0344, and TTHA1292) have been annotated in the HB8 genome, and electrophoretic mobility shift assays revealed that the TTHA0344 product interacts with the sip promoter region. In medium containing supersaturated silicic acids, free Fe(3+) levels were decreased due to Fe(3+) immobilization on colloidal silica. This suggests that, because Fe(3+) ions are captured by colloidal silica in geothermal water, Thermus cells are continuously exposed to the risk of iron deficiency. Considering that Sip is involved in iron acquisition, Sip production may be a strategy to survive under conditions of low iron availability in geothermal water.
Importance: The thermophilic bacterium Thermus thermophilus HB8 produces silica-induced protein (Sip) in the presence of supersaturated silicic acids. Sip has homology with iron-binding ABC transporter; however, the mechanism by which Sip expression is induced by silicic acids remains unexplained. We demonstrate that Sip captures iron and its transcription is regulated by the repressor ferric uptake regulator (Fur). This implies that Sip is expressed with iron deficiency. In addition, it is suggested that negatively charged colloidal silica in supersaturated solution absorbs Fe(3+) ions and decreases iron availability. Considering that geothermal water contains ample silicic acids, it is suggested that thermophilic bacteria are always facing iron starvation. Sip production may be a strategy for surviving under conditions of low iron availability in geothermal water.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Stimulation of expression of a silica-induced protein (Sip) in Thermus thermophilus by supersaturated silicic acid.Appl Environ Microbiol. 2009 Apr;75(8):2406-13. doi: 10.1128/AEM.02387-08. Epub 2009 Feb 20. Appl Environ Microbiol. 2009. PMID: 19233950 Free PMC article.
-
A tightly regulated expression system for E. coli using supersaturated silicic acid.Biotechnol Lett. 2016 Aug;38(8):1381-7. doi: 10.1007/s10529-016-2118-z. Epub 2016 May 5. Biotechnol Lett. 2016. PMID: 27146211
-
Silica deposition and phenotypic changes to Thermus thermophilus cultivated in the presence of supersaturated silicia.ISME J. 2010 Jun;4(6):809-16. doi: 10.1038/ismej.2010.12. Epub 2010 Mar 11. ISME J. 2010. PMID: 20220788
-
Molecular biology of S-layers.FEMS Microbiol Rev. 1997 Jun;20(1-2):47-98. doi: 10.1111/j.1574-6976.1997.tb00304.x. FEMS Microbiol Rev. 1997. PMID: 9276928 Review.
-
Type IV Pili in Thermophilic Bacteria: Mechanisms and Ecological Implications.Biomolecules. 2025 Mar 21;15(4):459. doi: 10.3390/biom15040459. Biomolecules. 2025. PMID: 40305182 Free PMC article. Review.
Cited by
-
Identification of the Preferred DNA-Binding Sequence and Transcription Regulatory Network for the Thermophilic Zinc Uptake Regulator TTHA1292.J Bacteriol. 2022 Nov 15;204(11):e0030322. doi: 10.1128/jb.00303-22. Epub 2022 Oct 26. J Bacteriol. 2022. PMID: 36286513 Free PMC article.
-
Development of a new gene expression vector for Thermus thermophilus using a silica-inducible promoter.Microb Cell Fact. 2020 Jun 8;19(1):126. doi: 10.1186/s12934-020-01385-2. Microb Cell Fact. 2020. PMID: 32513169 Free PMC article.
References
-
- Iler RK. 1979. The chemistry of silica. Wiley & Sons, New York, NY.
-
- Phoenix VR, Konhauser KO, Ferris FG. 2003. Experimental study of iron and silica immobilization by bacteria in mixed Fe-Si systems: implications for microbial silicification in hot springs. Can J Earth Sci 40:1669–1678. doi:10.1139/e03-044. - DOI
-
- Yee N, Phoenix VR, Konhauser KO, Benning LG, Ferris FG. 2003. The effect of cyanobacteria on silica precipitation at neutral pH: implications for bacterial silicification in geothermal hot springs. Chem Geol 199:83–90. doi:10.1016/S0009-2541(03)00120-7. - DOI
-
- Inagaki F, Hayashi S, Doi K, Motomura Y, Izawa E, Ogata S. 1997. Microbial participation in the formation of siliceous deposits from geothermal water and analysis of the extremely thermophilic bacterial community. FEMS Microbiol Ecol 24:41–48. doi:10.1111/j.1574-6941.1997.tb00421.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

