The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes
- PMID: 26996206
- PMCID: PMC4800398
- DOI: 10.1038/srep23121
The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes
Abstract
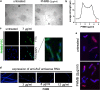
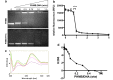
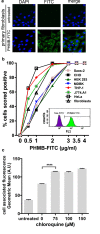
To combat infection and antimicrobial resistance, it is helpful to elucidate drug mechanism(s) of action. Here we examined how the widely used antimicrobial polyhexamethylene biguanide (PHMB) kills bacteria selectively over host cells. Contrary to the accepted model of microbial membrane disruption by PHMB, we observed cell entry into a range of bacterial species, and treated bacteria displayed cell division arrest and chromosome condensation, suggesting DNA binding as an alternative antimicrobial mechanism. A DNA-level mechanism was confirmed by observations that PHMB formed nanoparticles when mixed with isolated bacterial chromosomal DNA and its effects on growth were suppressed by pairwise combination with the DNA binding ligand Hoechst 33258. PHMB also entered mammalian cells, but was trapped within endosomes and excluded from nuclei. Therefore, PHMB displays differential access to bacterial and mammalian cellular DNA and selectively binds and condenses bacterial chromosomes. Because acquired resistance to PHMB has not been reported, selective chromosome condensation provides an unanticipated paradigm for antimicrobial action that may not succumb to resistance.
Conflict of interest statement
The authors declare competing interests. KC and LG are inventors on a filed patent application WO2013054123.
Figures



Similar articles
-
Bactericidal effects of polyhexamethylene biguanide against intracellular Staphylococcus aureus EMRSA-15 and USA 300.J Antimicrob Chemother. 2016 May;71(5):1252-9. doi: 10.1093/jac/dkv474. Epub 2016 Jan 28. J Antimicrob Chemother. 2016. PMID: 26825118
-
Effect of polyhexamethylene biguanide functionalized silver nanoparticles on the growth of Staphylococcus aureus.FEMS Microbiol Lett. 2019 Feb 1;366(4):fnz036. doi: 10.1093/femsle/fnz036. FEMS Microbiol Lett. 2019. PMID: 30879081
-
Polyvinyl Alcohol/Chitosan/Polyhexamethylene Biguanide Phase Separation System: A Potential Topical Antibacterial Formulation with Enhanced Antimicrobial Effect.Molecules. 2020 Mar 15;25(6):1334. doi: 10.3390/molecules25061334. Molecules. 2020. PMID: 32183411 Free PMC article.
-
Enhanced killing of Escherichia coli using a combination of polyhexamethylene biguanide hydrochloride and 1-bromo-3-chloro-5,5- dimethylimidazolidine-2,4-dione.FEMS Microbiol Lett. 2017 Nov 15;364(21). doi: 10.1093/femsle/fnx210. FEMS Microbiol Lett. 2017. PMID: 29029044
-
Polyhexamethylene biguanide and its antimicrobial role in wound healing: a narrative review.J Wound Care. 2023 Jan 2;32(1):5-20. doi: 10.12968/jowc.2023.32.1.5. J Wound Care. 2023. PMID: 36630111 Review.
Cited by
-
Microbial Community Profiling in Intensive Care Units Expose Limitations in Current Sanitary Standards.Front Public Health. 2019 Aug 28;7:240. doi: 10.3389/fpubh.2019.00240. eCollection 2019. Front Public Health. 2019. PMID: 31555629 Free PMC article.
-
Development of Antiviral CVC (Chief Value Cotton) Fabric.Polymers (Basel). 2021 Aug 5;13(16):2601. doi: 10.3390/polym13162601. Polymers (Basel). 2021. PMID: 34451140 Free PMC article.
-
Study on the Development of Antiviral Spandex Fabric Coated with Poly(hexamethylene biguanide) Hydrochloride (PHMB).Polymers (Basel). 2021 Jun 28;13(13):2122. doi: 10.3390/polym13132122. Polymers (Basel). 2021. PMID: 34203388 Free PMC article.
-
Synergism versus Additivity: Defining the Interactions between Common Disinfectants.mBio. 2021 Oct 26;12(5):e0228121. doi: 10.1128/mBio.02281-21. Epub 2021 Sep 21. mBio. 2021. PMID: 34544274 Free PMC article.
-
Recent Developments in Antimicrobial Polymers: A Review.Materials (Basel). 2016 Jul 20;9(7):599. doi: 10.3390/ma9070599. Materials (Basel). 2016. PMID: 28773721 Free PMC article. Review.
References
-
- Muller G. & Kramer A. Biocompatibility index of antiseptic agents by parallel assessment of antimicrobial activity and cellular cytotoxicity. J Antimicrob Chemother 61, 1281–1287 (2008). - PubMed
-
- Muller G., Koburger T. & Kramer A. Interaction of polyhexamethylene biguanide hydrochloride (PHMB) with phosphatidylcholine containing o/w emulsion and consequences for microbicidal efficacy and cytotoxicity. Chem Biol Interact 201, 58–64 (2013). - PubMed
-
- Wessels S. & Ingmer H. Modes of action of three disinfectant active substances: A review. Regul. Toxicol. Pharmacol. 67, 456–467 (2013). - PubMed
-
- Gilbert P., Das J. R., Jones M. V. & Allison D. G. Assessment of resistance towards biocides following the attachment of micro-organisms to, and growth on, surfaces. J. Appl. Microbiol. 91, 248–54 (2001). - PubMed
-
- Broxton P., Woodcock P. M. & Gilbert P. A study of the antibacterial activity of some polyhexamethylene biguanides towards Escherichia coli ATCC 8739. J Appl Bacteriol 54, 345–353 (1983). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

