Rhamnose synthase activity is required for pathogenicity of the vascular wilt fungus Verticillium dahliae
- PMID: 26996832
- PMCID: PMC6638212
- DOI: 10.1111/mpp.12401
Rhamnose synthase activity is required for pathogenicity of the vascular wilt fungus Verticillium dahliae
Abstract
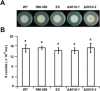
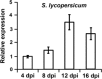
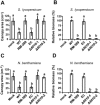
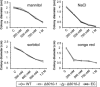
The initial interaction of a pathogenic fungus with its host is complex and involves numerous metabolic pathways and regulatory proteins. Considerable attention has been devoted to proteins that play a crucial role in these interactions, with an emphasis on so-called effector molecules that are secreted by the invading microbe to establish the symbiosis. However, the contribution of other types of molecules, such as glycans, is less well appreciated. Here, we present a random genetic screen that enabled us to identify 58 novel candidate genes that are involved in the pathogenic potential of the fungal pathogen Verticillium dahliae, which causes vascular wilt diseases in over 200 dicotyledonous plant species, including economically important crops. One of the candidate genes that was identified concerns a putative biosynthetic gene involved in nucleotide sugar precursor formation, as it encodes a putative nucleotide-rhamnose synthase/epimerase-reductase (NRS/ER). This enzyme has homology to bacterial enzymes involved in the biosynthesis of the nucleotide sugar deoxy-thymidine diphosphate (dTDP)-rhamnose, a precursor of L-rhamnose, which has been shown to be required for virulence in several human pathogenic bacteria. Rhamnose is known to be a minor cell wall glycan in fungi and has therefore not been suspected as a crucial molecule in fungal-host interactions. Nevertheless, our study shows that deletion of the VdNRS/ER gene from the V. dahliae genome results in complete loss of pathogenicity on tomato and Nicotiana benthamiana plants, whereas vegetative growth and sporulation are not affected. We demonstrate that VdNRS/ER is a functional enzyme in the biosynthesis of uridine diphosphate (UDP)-rhamnose, and further analysis has revealed that VdNRS/ER deletion strains are impaired in the colonization of tomato roots. Collectively, our results demonstrate that rhamnose, although only a minor cell wall component, is essential for the pathogenicity of V. dahliae.
Keywords: Agrobacterium tumefaciens-mediated transformation (ATMT); UDP-rhamnose; attachment; carbohydrate; root colonization; tomato; vascular wilt.
© 2016 BSPP AND JOHN WILEY & SONS LTD.
Figures



Similar articles
-
Identification of pathogenicity-related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens-mediated T-DNA insertional mutagenesis.Mol Biotechnol. 2011 Nov;49(3):209-21. doi: 10.1007/s12033-011-9392-8. Mol Biotechnol. 2011. PMID: 21424547 Free PMC article.
-
Genetic alteration of UDP-rhamnose metabolism in Botrytis cinerea leads to the accumulation of UDP-KDG that adversely affects development and pathogenicity.Mol Plant Pathol. 2017 Feb;18(2):263-275. doi: 10.1111/mpp.12398. Epub 2016 Jul 20. Mol Plant Pathol. 2017. PMID: 26991954 Free PMC article.
-
Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts.Mol Plant Pathol. 2017 May;18(4):596-608. doi: 10.1111/mpp.12520. Epub 2017 Feb 14. Mol Plant Pathol. 2017. PMID: 27911046 Free PMC article.
-
The Role of Pathogen-Secreted Proteins in Fungal Vascular Wilt Diseases.Int J Mol Sci. 2015 Oct 9;16(10):23970-93. doi: 10.3390/ijms161023970. Int J Mol Sci. 2015. PMID: 26473835 Free PMC article. Review.
-
Interactions between Verticillium dahliae and its host: vegetative growth, pathogenicity, plant immunity.Appl Microbiol Biotechnol. 2014 Aug;98(16):6921-32. doi: 10.1007/s00253-014-5863-8. Epub 2014 Jun 15. Appl Microbiol Biotechnol. 2014. PMID: 24928658 Review.
Cited by
-
VdOGDH is involved in energy metabolism and required for virulence of Verticillium dahliae.Curr Genet. 2020 Apr;66(2):345-359. doi: 10.1007/s00294-019-01025-2. Epub 2019 Aug 17. Curr Genet. 2020. PMID: 31422448
-
UDP-4-Keto-6-Deoxyglucose, a Transient Antifungal Metabolite, Weakens the Fungal Cell Wall Partly by Inhibition of UDP-Galactopyranose Mutase.mBio. 2017 Nov 21;8(6):e01559-17. doi: 10.1128/mBio.01559-17. mBio. 2017. PMID: 29162710 Free PMC article.
-
Genomic insights into Verticillium: a review of progress in the genomics era.Front Microbiol. 2024 Oct 11;15:1463779. doi: 10.3389/fmicb.2024.1463779. eCollection 2024. Front Microbiol. 2024. PMID: 39464398 Free PMC article. Review.
-
Diversity of Cell Wall Related Proteins in Human Pathogenic Fungi.J Fungi (Basel). 2017 Dec 29;4(1):6. doi: 10.3390/jof4010006. J Fungi (Basel). 2017. PMID: 29371499 Free PMC article.
-
The Nep1-like protein family of Magnaporthe oryzae is dispensable for the infection of rice plants.Sci Rep. 2017 Jun 29;7(1):4372. doi: 10.1038/s41598-017-04430-0. Sci Rep. 2017. PMID: 28663588 Free PMC article.
References
-
- Bolek, Y. , El‐Zik, K.M. , Pepper, A.E. , Bell, A.A. , Magill, C.W. , Thaxton, P. M. and Reddy, O. U. K. (2005) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590.
-
- Di Pietro, A. , Garcia‐MacEira, F.I. , Meglecz, E. and Roncero, M.I. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

