GTF2E2 Mutations Destabilize the General Transcription Factor Complex TFIIE in Individuals with DNA Repair-Proficient Trichothiodystrophy
- PMID: 26996949
- PMCID: PMC4833217
- DOI: 10.1016/j.ajhg.2016.02.008
GTF2E2 Mutations Destabilize the General Transcription Factor Complex TFIIE in Individuals with DNA Repair-Proficient Trichothiodystrophy
Abstract
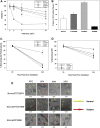
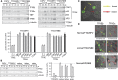
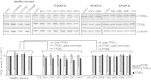
The general transcription factor IIE (TFIIE) is essential for transcription initiation by RNA polymerase II (RNA pol II) via direct interaction with the basal transcription/DNA repair factor IIH (TFIIH). TFIIH harbors mutations in two rare genetic disorders, the cancer-prone xeroderma pigmentosum (XP) and the cancer-free, multisystem developmental disorder trichothiodystrophy (TTD). The phenotypic complexity resulting from mutations affecting TFIIH has been attributed to the nucleotide excision repair (NER) defect as well as to impaired transcription. Here, we report two unrelated children showing clinical features typical of TTD who harbor different homozygous missense mutations in GTF2E2 (c.448G>C [p.Ala150Pro] and c.559G>T [p.Asp187Tyr]) encoding the beta subunit of transcription factor IIE (TFIIEβ). Repair of ultraviolet-induced DNA damage was normal in the GTF2E2 mutated cells, indicating that TFIIE was not involved in NER. We found decreased protein levels of the two TFIIE subunits (TFIIEα and TFIIEβ) as well as decreased phosphorylation of TFIIEα in cells from both children. Interestingly, decreased phosphorylation of TFIIEα was also seen in TTD cells with mutations in ERCC2, which encodes the XPD subunit of TFIIH, but not in XP cells with ERCC2 mutations. Our findings support the theory that TTD is caused by transcriptional impairments that are distinct from the NER disorder XP.
Copyright © 2016 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy.Hum Mutat. 2008 Oct;29(10):1194-208. doi: 10.1002/humu.20768. Hum Mutat. 2008. PMID: 18470933 Free PMC article.
-
Comparative study of nucleotide excision repair defects between XPD-mutated fibroblasts derived from trichothiodystrophy and xeroderma pigmentosum patients.DNA Repair (Amst). 2008 Dec 1;7(12):1990-8. doi: 10.1016/j.dnarep.2008.08.009. Epub 2008 Oct 10. DNA Repair (Amst). 2008. PMID: 18817897
-
TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription.Nat Commun. 2019 May 7;10(1):2084. doi: 10.1038/s41467-019-10131-1. Nat Commun. 2019. PMID: 31064989 Free PMC article.
-
XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase.DNA Repair (Amst). 2011 Jul 15;10(7):697-713. doi: 10.1016/j.dnarep.2011.04.028. Epub 2011 May 14. DNA Repair (Amst). 2011. PMID: 21571596 Free PMC article. Review.
-
A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor.DNA Repair (Amst). 2011 Jul 15;10(7):714-21. doi: 10.1016/j.dnarep.2011.04.021. Epub 2011 May 17. DNA Repair (Amst). 2011. PMID: 21592869 Review.
Cited by
-
Ribosomal Dysfunction Is a Common Pathomechanism in Different Forms of Trichothiodystrophy.Cells. 2023 Jul 17;12(14):1877. doi: 10.3390/cells12141877. Cells. 2023. PMID: 37508541 Free PMC article.
-
Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.Nat Struct Mol Biol. 2019 Jun;26(6):397-406. doi: 10.1038/s41594-019-0220-3. Epub 2019 May 20. Nat Struct Mol Biol. 2019. PMID: 31110295 Free PMC article.
-
Trans-acting genetic modifiers of clinical severity in heterozygous β-Thalassemia trait.Ann Hematol. 2024 Nov;103(11):4437-4447. doi: 10.1007/s00277-024-06007-0. Epub 2024 Sep 24. Ann Hematol. 2024. PMID: 39316111 Review.
-
TFIIH mutations can impact on translational fidelity of the ribosome.Hum Mol Genet. 2023 Mar 20;32(7):1102-1113. doi: 10.1093/hmg/ddac268. Hum Mol Genet. 2023. PMID: 36308430 Free PMC article.
-
C. elegans TFIIH subunit GTF-2H5/TTDA is a non-essential transcription factor indispensable for DNA repair.Commun Biol. 2021 Nov 25;4(1):1336. doi: 10.1038/s42003-021-02875-8. Commun Biol. 2021. PMID: 34824371 Free PMC article.
References
-
- Giglia-Mari G., Coin F., Ranish J.A., Hoogstraten D., Theil A., Wijgers N., Jaspers N.G., Raams A., Argentini M., van der Spek P.J. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004;36:714–719. - PubMed
-
- Nonnekens J., Perez-Fernandez J., Theil A.F., Gadal O., Bonnart C., Giglia-Mari G. Mutations in TFIIH causing trichothiodystrophy are responsible for defects in ribosomal RNA production and processing. Hum. Mol. Genet. 2013;22:2881–2893. - PubMed
-
- Compe E., Egly J.M. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012;13:343–354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

