Establishment of In Vitro FUS-Associated Familial Amyotrophic Lateral Sclerosis Model Using Human Induced Pluripotent Stem Cells
- PMID: 26997647
- PMCID: PMC4834049
- DOI: 10.1016/j.stemcr.2016.02.011
Establishment of In Vitro FUS-Associated Familial Amyotrophic Lateral Sclerosis Model Using Human Induced Pluripotent Stem Cells
Abstract
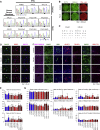
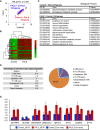
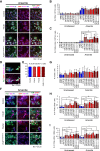
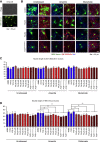
Amyotrophic lateral sclerosis (ALS) is a late-onset motor neuron disorder. Although its neuropathology is well understood, the cellular and molecular mechanisms are yet to be elucidated due to limitations in the currently available human genetic data. In this study, we generated induced pluripotent stem cells (iPSC) from two familial ALS (FALS) patients with a missense mutation in the fused-in sarcoma (FUS) gene carrying the heterozygous FUS H517D mutation, and isogenic iPSCs with the homozygous FUS H517D mutation by genome editing technology. These cell-derived motor neurons mimicked several neurodegenerative phenotypes including mis-localization of FUS into cytosolic and stress granules under stress conditions, and cellular vulnerability. Moreover, exon array analysis using motor neuron precursor cells (MPCs) combined with CLIP-seq datasets revealed aberrant gene expression and/or splicing pattern in FALS MPCs. These results suggest that iPSC-derived motor neurons are a useful tool for analyzing the pathogenesis of human motor neuron disorders.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Akiyama T., Warita H., Kato M., Nishiyama A., Izumi R., Ikeda C., Kamada M., Suzuki N., Aoki M. Genotype-phenotype relationships in familial ALS with FUS/TLS mutations in Japan. Muscle Nerve. 2016 - PubMed
-
- Almeida S., Gascon E., Tran H., Chou H.J., Gendron T.F., Degroot S., Tapper A.R., Sellier C., Charlet-Berguerand N., Karydas A. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. - PMC - PubMed
-
- Aruga J., Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol. Cell. Neurosci. 2003;24:117–129. - PubMed
-
- Atkin J.D., Farg M.A., Turner B.J., Tomas D., Lysaght J.A., Nunan J., Rembach A., Nagley P., Beart P.M., Cheema S.S. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J. Biol. Chem. 2006;281:30152–30165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

