Molecular Evolution and Spatial Transmission of Severe Fever with Thrombocytopenia Syndrome Virus Based on Complete Genome Sequences
- PMID: 26999664
- PMCID: PMC4801363
- DOI: 10.1371/journal.pone.0151677
Molecular Evolution and Spatial Transmission of Severe Fever with Thrombocytopenia Syndrome Virus Based on Complete Genome Sequences
Abstract
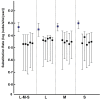
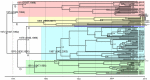
Severe fever with thrombocytopenia syndrome virus (SFTSV) was a novel tick-borne bunyavirus that caused hemorrhagic fever with a high fatality rate in East Asia. In this study we analyzed the complete genome sequences of 122 SFTSV strains to determine the phylogeny, evolution and reassortment of the virus. We revealed that the evolutionary rate of three genome segments were different, with highest in the S segment and lowest in the L segment. The SFTSV strains were phylogenetically classified into 5 lineages (A, B, C, D and E) with each genome segment. SFTSV strains from China were classified in all 5 lineages, strains from South Korea were classified into 3 lineages (A, D, and E), and all strains from Japan were classified in only linage E. Using the average evolutionary rate of the three genome segments, we found that the extant SFTSV originated 20-87 years ago in the Dabie Mountain area in central China. The viruses were then transmitted to other areas of China, Japan and South Korea. We also found that six SFTSV strains were reassortants. Selection pressure analysis suggested that SFTSV was under purifying selection according to the four genes (RNA-dependent RNA polymerase, glycoprotein, nucleocapsid protein, non-structural protein), and two sites (37, 1033) of glycoproteins were identified as being under strong positive selection. We concluded that SFTSV originated in central China and spread to other places recently and the virus was under purifying selection with high frequency of reassortment.
Conflict of interest statement
Figures

Similar articles
-
Molecular genomic characterization of tick- and human-derived severe fever with thrombocytopenia syndrome virus isolates from South Korea.PLoS Negl Trop Dis. 2017 Sep 22;11(9):e0005893. doi: 10.1371/journal.pntd.0005893. eCollection 2017 Sep. PLoS Negl Trop Dis. 2017. PMID: 28937979 Free PMC article.
-
Novel sub-lineages, recombinants and reassortants of severe fever with thrombocytopenia syndrome virus.Ticks Tick Borne Dis. 2017 Mar;8(3):385-390. doi: 10.1016/j.ttbdis.2016.12.015. Epub 2017 Jan 3. Ticks Tick Borne Dis. 2017. PMID: 28117273
-
Phylogenetic and Geographic Relationships of Severe Fever With Thrombocytopenia Syndrome Virus in China, South Korea, and Japan.J Infect Dis. 2015 Sep 15;212(6):889-98. doi: 10.1093/infdis/jiv144. Epub 2015 Mar 11. J Infect Dis. 2015. PMID: 25762790
-
Severe fever with thrombocytopenia syndrome and its pathogen SFTSV.Microbes Infect. 2015 Feb;17(2):149-54. doi: 10.1016/j.micinf.2014.12.002. Epub 2014 Dec 11. Microbes Infect. 2015. PMID: 25498868 Review.
-
[The molecular evolution of Dabie bandavirus (Phenuiviridae: Bandavirus: Dabie bandavirus), the agent of severe fever with thrombocytopenia syndrome].Vopr Virusol. 2022 Jan 8;66(6):409-416. doi: 10.36233/0507-4088-68. Vopr Virusol. 2022. PMID: 35019247 Review. Russian.
Cited by
-
Advancements in the Worldwide Detection of Severe Fever with Thrombocytopenia Syndrome Virus Infection from 2009 to 2023.China CDC Wkly. 2023 Aug 4;5(31):687-693. doi: 10.46234/ccdcw2023.132. China CDC Wkly. 2023. PMID: 37593140 Free PMC article.
-
Detection and prevalence of a novel Bandavirus related to Guertu virus in Amblyomma gemma ticks and human populations in Isiolo County, Kenya.PLoS One. 2024 Sep 20;19(9):e0310862. doi: 10.1371/journal.pone.0310862. eCollection 2024. PLoS One. 2024. PMID: 39302958 Free PMC article.
-
Current status of severe fever with thrombocytopenia syndrome in China.Virol Sin. 2017 Feb;32(1):51-62. doi: 10.1007/s12250-016-3931-1. Epub 2017 Feb 27. Virol Sin. 2017. PMID: 28251515 Free PMC article. Review.
-
Analysis of Gene Differences Between F and B Epidemic Lineages of Bandavirus Dabieense.Microorganisms. 2025 Jan 28;13(2):292. doi: 10.3390/microorganisms13020292. Microorganisms. 2025. PMID: 40005658 Free PMC article.
-
Baseline mapping of severe fever with thrombocytopenia syndrome virology, epidemiology and vaccine research and development.NPJ Vaccines. 2020 Dec 17;5(1):111. doi: 10.1038/s41541-020-00257-5. NPJ Vaccines. 2020. PMID: 33335100 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

