Potential Sources and Transmission of Salmonella and Antimicrobial Resistance in Kampala, Uganda
- PMID: 26999788
- PMCID: PMC4801205
- DOI: 10.1371/journal.pone.0152130
Potential Sources and Transmission of Salmonella and Antimicrobial Resistance in Kampala, Uganda
Abstract
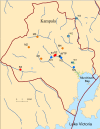
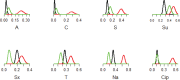
In sub‒Saharan Africa, non‒typhoidal Salmonellae (NTS) cause invasive disease particularly in children and HIV infected adults, but the disease epidemiology is poorly understood. Between 2012 and 2013, we investigated NTS sources and transmission in Kampala. We detected Salmonella in 60% of the influent and 60% of the effluent samples from a wastewater treatment plant and 53.3% of the influent and 10% of the effluent samples from waste stabilization ponds that serve the human population; 40.9% of flush‒water samples from ruminant slaughterhouses, 6.6% of the poultry fecal samples from live bird markets and 4% of the fecal samples from swine at slaughter; and in 54.2% of the water samples from a channel that drains storm-water and effluents from the city. We obtained 775 Salmonella isolates, identified 32 serovars, and determined resistance to 15 antimicrobials. We genotyped common serovars using multiple‒locus variable number tandem repeats analysis or pulsed‒field gel electrophoresis. In addition, we analyzed 49 archived NTS isolates from asymptomatic livestock and human clinical cases. Salmonella from ruminant and swine sources were mostly pan‒susceptible (95%) while poultry isolates were generally more resistant. Salmonella Kentucky isolated from poultry exhibited extensive drug resistance characterized by resistance to 10 antimicrobials. Interestingly, similar genotypes of S. Kentucky but with less antimicrobial resistance (AMR) were found in poultry, human and environmental sources. The observed AMR patterns could be attributed to host or management factors associated with production. Alternatively, S. Kentucky may be prone to acquiring AMR. The factors driving AMR remain poorly understood and should be elucidated. Overall, shared genotypes and AMR phenotypes were found in NTS from human, livestock and environmental sources, suggesting zoonotic and environmental transmissions most likely occur. Information from this study could be used to control NTS transmission.
Conflict of interest statement
Figures


References
-
- Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, et al. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin Infect Dis 2012; 54: 472–479. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

