Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea
- PMID: 27001046
- PMCID: PMC4838380
- DOI: 10.1093/nar/gkw153
Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea
Abstract
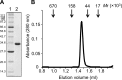
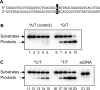
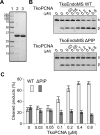
The common mismatch repair system processed by MutS and MutL and their homologs was identified in Bacteria and Eukarya. However, no evidence of a functional MutS/L homolog has been reported for archaeal organisms, and it is not known whether the mismatch repair system is conserved in Archaea. Here, we describe an endonuclease that cleaves double-stranded DNA containing a mismatched base pair, from the hyperthermophilic archaeon Pyrococcus furiosus The corresponding gene revealed that the activity originates from PF0012, and we named this enzyme Endonuclease MS (EndoMS) as the mismatch-specific Endonuclease. The sequence similarity suggested that EndoMS is the ortholog of NucS isolated from Pyrococcus abyssi, published previously. Biochemical characterizations of the EndoMS homolog from Thermococcus kodakarensis clearly showed that EndoMS specifically cleaves both strands of double-stranded DNA into 5'-protruding forms, with the mismatched base pair in the central position. EndoMS cleaves G/T, G/G, T/T, T/C and A/G mismatches, with a more preference for G/T, G/G and T/T, but has very little or no effect on C/C, A/C and A/A mismatches. The discovery of this endonuclease suggests the existence of a novel mismatch repair process, initiated by the double-strand break generated by the EndoMS endonuclease, in Archaea and some Bacteria.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Barnes D.E., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. - PubMed
-
- Reardon J.T., Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. - PubMed
-
- Iyer R.R., Pluciennik A., Burdett V., Modrich P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

