Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer
- PMID: 27001519
- PMCID: PMC4872098
- DOI: 10.1093/nar/gkw177
Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer
Abstract
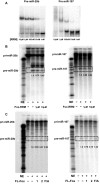
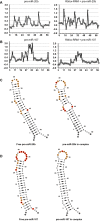
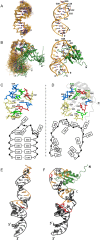
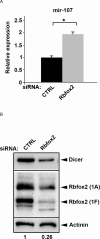
Rbfox proteins regulate tissue-specific splicing by targeting a conserved GCAUG sequence within pre-mRNAs. We report here that sequence-specific binding of the conserved Rbfox RRM to miRNA precursors containing the same sequence motif in their terminal loops, including miR-20b and miR-107, suppresses their nuclear processing. The structure of the complex between precursor miR-20b and Rbfox RRM shows the molecular basis for recognition, and reveals changes in the stem-loop upon protein binding. In mammalian cells, Rbfox2 downregulates mature miR-20b and miR-107 levels and increases the expression of their downstream targets PTEN and Dicer, respectively, suggesting that Rbfox2 indirectly regulates many more cellular miRNAs. Thus, some of the widespread cellular functions of Rbfox2 protein are attributable to regulation of miRNA biogenesis, and might include the mis-regulation of miR-20b and miR-107 in cancer and neurodegeneration.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis.Mol Cell. 2017 Apr 20;66(2):270-284.e13. doi: 10.1016/j.molcel.2017.03.014. Mol Cell. 2017. PMID: 28431233
-
MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation.Mol Cell. 2011 Nov 4;44(3):424-36. doi: 10.1016/j.molcel.2011.09.012. Mol Cell. 2011. PMID: 22055188
-
Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):E1881-9. doi: 10.1073/pnas.1602532113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976605 Free PMC article.
-
Mechanisms of control of microRNA biogenesis.J Biochem. 2010 Oct;148(4):381-92. doi: 10.1093/jb/mvq096. Epub 2010 Sep 9. J Biochem. 2010. PMID: 20833630 Free PMC article. Review.
-
Regulation of microRNA biogenesis and function.Thromb Haemost. 2012 Apr;107(4):605-10. doi: 10.1160/TH11-12-0836. Epub 2012 Feb 8. Thromb Haemost. 2012. PMID: 22318703 Review.
Cited by
-
Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity.Nat Commun. 2023 Feb 9;14(1):701. doi: 10.1038/s41467-023-36394-3. Nat Commun. 2023. PMID: 36759600 Free PMC article.
-
Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.Nat Commun. 2018 Jun 26;9(1):2479. doi: 10.1038/s41467-018-04871-9. Nat Commun. 2018. PMID: 29946118 Free PMC article.
-
Stress Granules Contain Rbfox2 with Cell Cycle-related mRNAs.Sci Rep. 2017 Sep 11;7(1):11211. doi: 10.1038/s41598-017-11651-w. Sci Rep. 2017. PMID: 28894257 Free PMC article.
-
RNA-binding proteins regulating the CD44 alternative splicing.Front Mol Biosci. 2023 Dec 1;10:1326148. doi: 10.3389/fmolb.2023.1326148. eCollection 2023. Front Mol Biosci. 2023. PMID: 38106992 Free PMC article. Review.
-
Differentially Expressed miRNAs Influence Metabolic Processes in Pituitary Oncocytoma.Neurochem Res. 2019 Oct;44(10):2360-2371. doi: 10.1007/s11064-019-02789-2. Epub 2019 Apr 3. Neurochem Res. 2019. PMID: 30945144 Free PMC article.
References
-
- Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. - PubMed
-
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

