Functional annotation of the vlinc class of non-coding RNAs using systems biology approach
- PMID: 27001520
- PMCID: PMC4838384
- DOI: 10.1093/nar/gkw162
Functional annotation of the vlinc class of non-coding RNAs using systems biology approach
Abstract
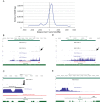
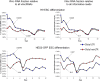
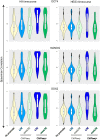
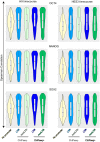
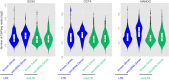
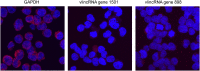
Functionality of the non-coding transcripts encoded by the human genome is the coveted goal of the modern genomics research. While commonly relied on the classical methods of forward genetics, integration of different genomics datasets in a global Systems Biology fashion presents a more productive avenue of achieving this very complex aim. Here we report application of a Systems Biology-based approach to dissect functionality of a newly identified vast class of very long intergenic non-coding (vlinc) RNAs. Using highly quantitative FANTOM5 CAGE dataset, we show that these RNAs could be grouped into 1542 novel human genes based on analysis of insulators that we show here indeed function as genomic barrier elements. We show that vlinc RNAs genes likely function in cisto activate nearby genes. This effect while most pronounced in closely spaced vlinc RNA-gene pairs can be detected over relatively large genomic distances. Furthermore, we identified 101 vlinc RNA genes likely involved in early embryogenesis based on patterns of their expression and regulation. We also found another 109 such genes potentially involved in cellular functions also happening at early stages of development such as proliferation, migration and apoptosis. Overall, we show that Systems Biology-based methods have great promise for functional annotation of non-coding RNAs.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Herbert A., Rich A. RNA processing in evolution. The logic of soft-wired genomes. Ann. N Y Acad. Sci. 1999;870:119–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

