Low-Frequency Drug Resistance in HIV-Infected Ugandans on Antiretroviral Treatment Is Associated with Regimen Failure
- PMID: 27001818
- PMCID: PMC4879357
- DOI: 10.1128/AAC.00038-16
Low-Frequency Drug Resistance in HIV-Infected Ugandans on Antiretroviral Treatment Is Associated with Regimen Failure
Abstract
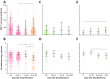
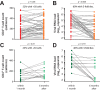
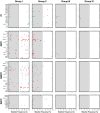
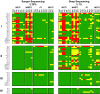
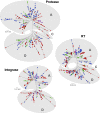
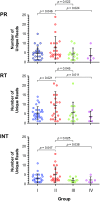
Most patients failing antiretroviral treatment in Uganda continue to fail their treatment regimen even if a dominant drug-resistant HIV-1 genotype is not detected. In a recent retrospective study, we observed that approximately 30% of HIV-infected individuals in the Joint Clinical Research Centre (Kampala, Uganda) experienced virologic failure with a susceptible HIV-1 genotype based on standard Sanger sequencing. Selection of minority drug-resistant HIV-1 variants (not detectable by Sanger sequencing) under antiretroviral therapy pressure can lead to a shift in the viral quasispecies distribution, becoming dominant members of the virus population and eventually causing treatment failure. Here, we used a novel HIV-1 genotyping assay based on deep sequencing (DeepGen) to quantify low-level drug-resistant HIV-1 variants in 33 patients failing a first-line antiretroviral treatment regimen in the absence of drug-resistant mutations, as screened by standard population-based Sanger sequencing. Using this sensitive assay, we observed that 64% (21/33) of these individuals had low-frequency (or minority) drug-resistant variants in the intrapatient HIV-1 population, which correlated with treatment failure. Moreover, the presence of these minority HIV-1 variants was associated with higher intrapatient HIV-1 diversity, suggesting a dynamic selection or fading of drug-resistant HIV-1 variants from the viral quasispecies in the presence or absence of drug pressure, respectively. This study identified low-frequency HIV drug resistance mutations by deep sequencing in Ugandan patients failing antiretroviral treatment but lacking dominant drug resistance mutations as determined by Sanger sequencing methods. We showed that these low-abundance drug-resistant viruses could have significant consequences for clinical outcomes, especially if treatment is not modified based on a susceptible HIV-1 genotype by Sanger sequencing. Therefore, we propose to make clinical decisions using more sensitive methods to detect minority HIV-1 variants.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- UNAIDS. 2013. Global update on HIV treatment 2013: results, impact, and opportunities. UNAIDS, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/85327/1/WHO_HIV_2013.9_eng.pdf.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

