Neural basis of reward anticipation and its genetic determinants
- PMID: 27001827
- PMCID: PMC4833244
- DOI: 10.1073/pnas.1503252113
Neural basis of reward anticipation and its genetic determinants
Abstract
Dysfunctional reward processing is implicated in various mental disorders, including attention deficit hyperactivity disorder (ADHD) and addictions. Such impairments might involve different components of the reward process, including brain activity during reward anticipation. We examined brain nodes engaged by reward anticipation in 1,544 adolescents and identified a network containing a core striatal node and cortical nodes facilitating outcome prediction and response preparation. Distinct nodes and functional connections were preferentially associated with either adolescent hyperactivity or alcohol consumption, thus conveying specificity of reward processing to clinically relevant behavior. We observed associations between the striatal node, hyperactivity, and the vacuolar protein sorting-associated protein 4A (VPS4A) gene in humans, and the causal role of Vps4 for hyperactivity was validated in Drosophila Our data provide a neurobehavioral model explaining the heterogeneity of reward-related behaviors and generate a hypothesis accounting for their enduring nature.
Keywords: GWAS; VPS4A; dopamine receptor; fMRI; neural network.
Conflict of interest statement
Conflict of interest statement: T.B. has served as an adviser or consultant to Eli Lilly, Hexal Pharma, Medice, Novartis, Otsuka, Oxford Outcomes, PCM Scientific, Shire, and Vifor Pharma; he has received conference attendance support, conference support, or speaking fees from Eli Lilly, Medice, Novartis, and Shire, and he is involved in clinical trials conducted by Eli Lilly, Shire, and Vifor Pharma. G.J.B. has received honoraria for teaching from GE Healthcare and has served as a consultant for IXICO. A.L.W.B. has received funding from Science Foundation Ireland. T.R. has served as a consultant for Cambridge Cognition, Eli Lilly, Lundbeck, Otsuka, Shire, and Teva; he has received research support from Eli Lilly, GlaxoSmithKline, and Lundbeck; editorial honoraria from Elsevier and Springer-Verlag; educational speaking honoraria from Merck, Sharp, and Dohme; and royalties from Cambridge Cognition. The other authors report no financial relationships with commercial interests.
Figures




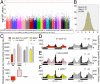
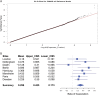

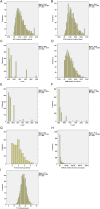
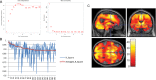
References
-
- Knutson B, Wimmer GE. Splitting the difference: How does the brain code reward episodes? Ann N Y Acad Sci. 2007;1104:54–69. - PubMed
-
- Nymberg C, et al. IMAGEN Consortium Neural mechanisms of attention-deficit/hyperactivity disorder symptoms are stratified by MAOA genotype. Biol Psychiatry. 2013;74(8):607–614. - PubMed
-
- Peters J, et al. IMAGEN Consortium Lower ventral striatal activation during reward anticipation in adolescent smokers. Am J Psychiatry. 2011;168(5):540–549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AA019526/AA/NIAAA NIH HHS/United States
- G0901858/MRC_/Medical Research Council/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
- R21 AA022404/AA/NIAAA NIH HHS/United States
- T32 DA007290/DA/NIDA NIH HHS/United States
- R01 MH085772/MH/NIMH NIH HHS/United States
- 93558/MRC_/Medical Research Council/United Kingdom
- T32 NS007413/NS/NINDS NIH HHS/United States
- MR/N000390/1/MRC_/Medical Research Council/United Kingdom
- 102215/WT_/Wellcome Trust/United Kingdom
- U54 EB020403/EB/NIBIB NIH HHS/United States
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

