MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression
- PMID: 27001906
- PMCID: PMC4804167
- DOI: 10.1038/ncomms10993
MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression
Abstract
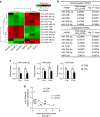
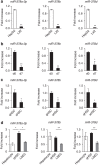
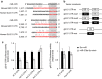
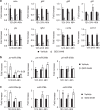
Hedgehog (Hh) signalling regulates hepatic fibrogenesis. MicroRNAs (miRNAs) mediate various cellular processes; however, their role in liver fibrosis is unclear. Here we investigate regulation of miRNAs in chronically damaged fibrotic liver. MiRNA profiling shows that expression of miR-378 family members (miR-378a-3p, miR-378b and miR-378d) declines in carbon tetrachloride (CCl4)-treated compared with corn-oil-treated mice. Overexpression of miR-378a-3p, directly targeting Gli3 in activated hepatic stellate cells (HSCs), reduces expression of Gli3 and profibrotic genes but induces gfap, the inactivation marker of HSCs, in CCl4-treated liver. Smo blocks transcriptional expression of miR-378a-3p by activating the p65 subunit of nuclear factor-κB (NF-κB). The hepatic level of miR-378a-3p is inversely correlated with the expression of Gli3 in tumour and non-tumour tissues in human hepatocellular carcinoma. Our results demonstrate that miR-378a-3p suppresses activation of HSCs by targeting Gli3 and its expression is regulated by Smo-dependent NF-κB signalling, suggesting miR-378a-3p has therapeutic potential for liver fibrosis.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

