Suppression of WHITE COLLAR-independent frequency Transcription by Histone H3 Lysine 36 Methyltransferase SET-2 Is Necessary for Clock Function in Neurospora
- PMID: 27002152
- PMCID: PMC4900255
- DOI: 10.1074/jbc.M115.711333
Suppression of WHITE COLLAR-independent frequency Transcription by Histone H3 Lysine 36 Methyltransferase SET-2 Is Necessary for Clock Function in Neurospora
Abstract
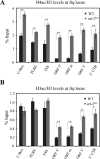
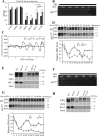
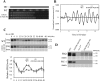
The circadian system in Neurospora is based on the transcriptional/translational feedback loops and rhythmic frequency (frq) transcription requires the WHITE COLLAR (WC) complex. Our previous paper has shown that frq could be transcribed in a WC-independent pathway in a strain lacking the histone H3K36 methyltransferase, SET-2 (su(var)3-9-enhancer-of-zeste-trithorax-2) (1), but the mechanism was unclear. Here we disclose that loss of histone H3K36 methylation, due to either deletion of SET-2 or H3K36R mutation, results in arrhythmic frq transcription and loss of overt rhythmicity. Histone acetylation at frq locus increases in set-2(KO) mutant. Consistent with these results, loss of H3K36 methylation readers, histone deacetylase RPD-3 (reduced potassium dependence 3) or EAF-3 (essential SAS-related acetyltransferase-associated factor 3), also leads to hyperacetylation of histone at frq locus and WC-independent frq expression, suggesting that proper chromatin modification at frq locus is required for circadian clock operation. Furthermore, a mutant strain with three amino acid substitutions (histone H3 lysine 9, 14, and 18 to glutamine) was generated to mimic the strain with hyperacetylation state of histone H3. H3K9QK14QK18Q mutant exhibits the same defective clock phenotype as rpd-3(KO) mutant. Our results support a scenario in which H3K36 methylation is required to establish a permissive chromatin state for circadian frq transcription by maintaining proper acetylation status at frq locus.
Keywords: Neurospora; SET-2 pathway; WC-independent frq; circadian rhythm; clock gene; gene transcription; histone modification.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

