Sensitive Visual Detection of AHPND Bacteria Using Loop-Mediated Isothermal Amplification Combined with DNA-Functionalized Gold Nanoparticles as Probes
- PMID: 27003504
- PMCID: PMC4803327
- DOI: 10.1371/journal.pone.0151769
Sensitive Visual Detection of AHPND Bacteria Using Loop-Mediated Isothermal Amplification Combined with DNA-Functionalized Gold Nanoparticles as Probes
Retraction in
-
Retraction: Sensitive Visual Detection of AHPND Bacteria Using Loop-Mediated Isothermal Amplification Combined with DNA-Functionalized Gold Nanoparticles as Probes.PLoS One. 2023 Feb 24;18(2):e0282453. doi: 10.1371/journal.pone.0282453. eCollection 2023. PLoS One. 2023. PMID: 36827257 Free PMC article. No abstract available.
Abstract
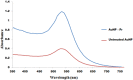
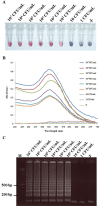
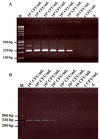
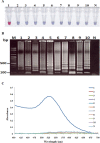
Acute hepatopancreatic necrosis disease (AHPND) is a component cause of early mortality syndrome (EMS) of shrimp. In 2013, the causative agent was found to be unique isolates of Vibrio parahaemolyticus (VPAHPND) that contained a 69 kbp plasmid (pAP1) carrying binary Pir-like toxin genes PirvpA and PirvpB. In Thailand, AHPND was first recognized in 2012, prior to knowledge of the causative agent, and it subsequently led to a precipitous drop in shrimp production. After VPAHPND was characterized, a major focus of the AHPND control strategy was to monitor broodstock shrimp and post larvae for freedom from VPAHPND by nucleic acid amplification methods, most of which required use of expensive and sophisticated equipment not readily available in a shrimp farm setting. Here, we describe a simpler but equally sensitive approach for detection of VPAHPND based on loop-mediated isothermal amplification (LAMP) combined with unaided visual reading of positive amplification products using a DNA-functionalized, ssDNA-labled nanogold probe (AuNP). The target for the special set of six LAMP primers used was the VPAHPND PirvpA gene. The LAMP reaction was carried out at 65°C for 45 min followed by addition of the red AuNP solution and further incubation at 65°C for 5 min, allowing any PirvpA gene amplicons present to hybridize with the probe. Hybridization protected the AuNP against aggregation, so that the solution color remained red upon subsequent salt addition (positive test result) while unprotected AuNP aggregated and underwent a color change from red to blue and eventually precipitated (negative result). The total assay time was approximately 50 min. The detection limit (100 CFU) was comparable to that of other commonly-used methods for nested PCR detection of VPAHPND and 100-times more sensitive than 1-step PCR detection methods (104 CFU) that used amplicon detection by electrophoresis or spectrophotometry. There was no cross reaction with DNA templates derived from non-AHPND bacteria commonly found in shrimp ponds (including other Vibrio species). The new method significantly reduced the time, difficulty and cost for molecular detection of VPAHPND in shrimp hatchery and farm settings.
Conflict of interest statement
Figures



Similar articles
-
Characterization and PCR Detection Of Binary, Pir-Like Toxins from Vibrio parahaemolyticus Isolates that Cause Acute Hepatopancreatic Necrosis Disease (AHPND) in Shrimp.PLoS One. 2015 May 27;10(5):e0126987. doi: 10.1371/journal.pone.0126987. eCollection 2015. PLoS One. 2015. PMID: 26017673 Free PMC article.
-
Loop-mediated isothermal amplification combined with colorimetric nanogold for detection of the microsporidian Enterocytozoon hepatopenaei in penaeid shrimp.J Appl Microbiol. 2013 May;114(5):1254-63. doi: 10.1111/jam.12160. Epub 2013 Mar 6. J Appl Microbiol. 2013. Retraction in: J Appl Microbiol. 2023 Mar 1;134(3):lxad027. doi: 10.1093/jambio/lxad027. PMID: 23387348 Retracted.
-
A Natural Vibrio parahaemolyticus ΔpirA Vp pirB Vp+ Mutant Kills Shrimp but Produces neither Pir Vp Toxins nor Acute Hepatopancreatic Necrosis Disease Lesions.Appl Environ Microbiol. 2017 Aug 1;83(16):e00680-17. doi: 10.1128/AEM.00680-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28576761 Free PMC article.
-
Rapid and sensitive detection of shrimp yellow head virus by loop-mediated isothermal amplification combined with a lateral flow dipstick.J Virol Methods. 2013 Mar;188(1-2):51-6. doi: 10.1016/j.jviromet.2012.11.041. Epub 2012 Dec 7. J Virol Methods. 2013. PMID: 23219929 Review.
-
VPAHPND pathogenesis and its mediated immune response in shrimp.Dev Comp Immunol. 2025 Jul 21;170:105429. doi: 10.1016/j.dci.2025.105429. Online ahead of print. Dev Comp Immunol. 2025. PMID: 40701311 Review.
Cited by
-
Multifuntional Gold Nanoparticles for the SERS Detection of Pathogens Combined with a LAMP-in-Microdroplets Approach.Materials (Basel). 2020 Apr 20;13(8):1934. doi: 10.3390/ma13081934. Materials (Basel). 2020. PMID: 32325992 Free PMC article.
-
Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods.Front Mol Biosci. 2021 Apr 20;8:637559. doi: 10.3389/fmolb.2021.637559. eCollection 2021. Front Mol Biosci. 2021. PMID: 33959631 Free PMC article. Review.
-
Combining Direct PCR Technology and Capillary Electrophoresis for an Easy-to-Operate and Highly Sensitive Infectious Disease Detection System for Shrimp.Life (Basel). 2022 Feb 13;12(2):276. doi: 10.3390/life12020276. Life (Basel). 2022. PMID: 35207563 Free PMC article.
-
Sensitivity Validation of EWOD Devices for Diagnosis of Early Mortality Syndrome (EMS) in Shrimp Using Colorimetric LAMP-XO Technique.Sensors (Basel). 2021 Apr 30;21(9):3126. doi: 10.3390/s21093126. Sensors (Basel). 2021. PMID: 33946302 Free PMC article.
-
Integration of RT-LAMP and Microfluidic Technology for Detection of SARS-CoV-2 in Wastewater as an Advanced Point-of-Care Platform.Food Environ Virol. 2022 Dec;14(4):364-373. doi: 10.1007/s12560-022-09522-3. Epub 2022 May 4. Food Environ Virol. 2022. PMID: 35508752 Free PMC article.
References
-
- Leano EM, Mohan CV. Early mortality syndrome threatens Asia’s shrimp farms. Glob Aquacult Advocate, Jul-Aug 2012: 38–39.
-
- Yang YT, Chen IT, Lee CT, Chen CY, Lin SS, Hor LI, et al. Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc. 2014; 2(5): e00816–14. 10.1128/genomeA.00816-14 - DOI - PMC - PubMed
-
- Sirikharin R, Taengchaiyaphum S, Sanguanrut P, Chi TD, Mavichak R, Proespraiwong P, et al. Characterization and PCR Detection of binary, pir-like toxins from Vibrio parahaemolyticus isolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp. PLoS One. 2015; 10: e0126987 10.1371/journal.pone.0126987 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

