The Eukaryotic Replisome Goes Under the Microscope
- PMID: 27003891
- PMCID: PMC4859309
- DOI: 10.1016/j.cub.2016.02.034
The Eukaryotic Replisome Goes Under the Microscope
Abstract
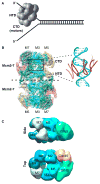
The machinery at the eukaryotic replication fork has seen many new structural advances using electron microscopy and crystallography. Recent structures of eukaryotic replisome components include the Mcm2-7 complex, the CMG helicase, DNA polymerases, a Ctf4 trimer hub and the first look at a core replisome of 20 different proteins containing the helicase, primase, leading polymerase and a lagging strand polymerase. The eukaryotic core replisome shows an unanticipated architecture, with one polymerase sitting above the helicase and the other below. Additionally, structures of Mcm2 bound to an H3/H4 tetramer suggest a direct role of the replisome in handling nucleosomes, which are important to DNA organization and gene regulation. This review provides a summary of some of the many recent advances in the structure of the eukaryotic replisome.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures



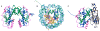
References
-
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. - PubMed
-
- Kornberg A, Baker TA. DNA Replication. New York: W.H. Freeman; 1992.
-
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. - PubMed
-
- Yao NY, O’Donnell ME. Comparison of bacterial and eukaryotic replisome components. In: Bradshaw RA, Stahl PD, editors. Encyclopedia of Cell Biology. Academic Press; 2015. pp. 396–417.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

