Genomic landscape of DNA repair genes in cancer
- PMID: 27004405
- PMCID: PMC5029628
- DOI: 10.18632/oncotarget.8196
Genomic landscape of DNA repair genes in cancer
Abstract
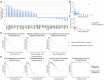
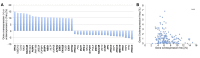
DNA repair genes are frequently mutated in cancer, yet limited data exist regarding the overall genomic landscape and functional implications of these alterations in their entirety. We created comprehensive lists of DNA repair genes and indirect caretakers. Mutation, copy number variation (CNV), and expression frequencies of these genes were analyzed in COSMIC. Mutation co-occurrence, clinical outcomes, and mutation burden were analyzed in TCGA. We report the 20 genes most frequently with mutations (n > 19,689 tumor samples for each gene), CNVs (n > 1,556), or up- or down-regulated (n = 7,998). Mutual exclusivity was observed as no genes displayed both high CNV gain and loss or high up- and down-regulation, and CNV gain and loss positively correlated with up- and down-regulation, respectively. Co-occurrence of mutations differed between cancers, and mutations in many DNA repair genes were associated with higher total mutation burden. Mutation and CNV frequencies offer insights into which genes may play tumor suppressive or oncogenic roles, such as NEIL2 and RRM2B, respectively. Mutual exclusivities within CNV and expression frequencies, and correlations between CNV and expression, support the functionality of these genomic alterations. This study provides comprehensive lists of candidate genes as potential biomarkers for genomic instability, novel therapeutic targets, or predictors of immunotherapy efficacy.
Keywords: DNA repair; cancer; copy number variations; gene expression; mutations.
Conflict of interest statement
None.
Figures



References
-
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. - PubMed
-
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nature reviews Cancer. 2012;12:801–817. - PubMed
-
- Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends in genetics. 2014;30:326–339. - PubMed
-
- Petrucelli N, Daly MB, Feldman GL. BRCA1 and BRCA2 Hereditary Breast and Ovarian Cancer. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1993.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

