Imprinted genes and imprinting control regions show predominant intermediate methylation in adult somatic tissues
- PMID: 27004446
- PMCID: PMC5066126
- DOI: 10.2217/epi.16.8
Imprinted genes and imprinting control regions show predominant intermediate methylation in adult somatic tissues
Abstract
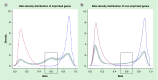
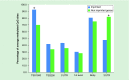
Genomic imprinting is an epigenetic feature characterized by parent-specific monoallelic gene expression. The aim of this study was to compare the DNA methylation status of imprinted genes and imprinting control regions (ICRs), harboring differentially methylated regions (DMRs) in a comprehensive panel of 18 somatic tissues. The germline DMRs analyzed were divided into ubiquitously imprinted and placenta-specific DMRs, which show identical and different methylation imprints in adult somatic and placental tissues, respectively. We showed that imprinted genes and ICR DMRs maintain methylation patterns characterized by intermediate methylation levels in somatic tissues, which are pronounced in a specific region of the promoter area, located 200-1500 bp from the transcription start site. This intermediate methylation is concordant with gene expression from a single unmethylated allele and silencing of a reciprocal parental allele through DNA methylation. The only exceptions were seen for ICR DMRs of placenta-specific imprinted genes, which showed low levels of methylation, suggesting that these genes escape parent-specific epigenetic regulation in somatic tissues.
Keywords: ICR; Levene's test; genomic imprinting; methylation; somatic tissues.
Conflict of interest statement
Financial & competing interests disclosure This research was funded by grants from the European Union's Horizon 2020 research grants ePerMed and WIDENLIFE; innovation programme (grant 692065); Enterprise Estonia (grant EU30020 and EU48695); Estonian Ministry of Education and Research (grant IUT34-16); EU-FP7 Eurostars Program (grant NOTED, EU41564); EU-FP7 IAPP Project (grant SARM, EU324509); Estonian Research Council (grants IUT20-60 and PUT736); the Development Fund of the University of Tartu (grant SP1GVARENG); EU structural support through Archimedes Foundation (grant 3.2.1001.11-0033); the European Regional Development Fund to the Centre of Excellence in Genomics (EXCEGEN; grant 3.2.0304.11-0312) and Wellcome Trust Senior Fellowship in Basic Biomedical Science (grant WT098017). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Figures

References
-
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–183. - PubMed
-
- Surani MA, Barton SC. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science. 1983;222(4627):1034–1036. - PubMed
-
- Lindsay RS, Kobes S, Knowler WC, Hanson RL. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of birth weight. Hum. Genet. 2002;110(5):503–509. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
