ABC-F Proteins Mediate Antibiotic Resistance through Ribosomal Protection
- PMID: 27006457
- PMCID: PMC4807367
- DOI: 10.1128/mBio.01975-15
ABC-F Proteins Mediate Antibiotic Resistance through Ribosomal Protection
Abstract
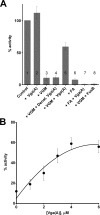
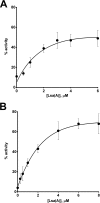
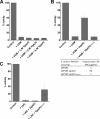
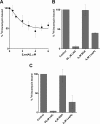
Members of the ABC-F subfamily of ATP-binding cassette proteins mediate resistance to a broad array of clinically important antibiotic classes that target the ribosome of Gram-positive pathogens. The mechanism by which these proteins act has been a subject of long-standing controversy, with two competing hypotheses each having gained considerable support: antibiotic efflux versus ribosomal protection. Here, we report on studies employing a combination of bacteriological and biochemical techniques to unravel the mechanism of resistance of these proteins, and provide several lines of evidence that together offer clear support to the ribosomal protection hypothesis. Of particular note, we show that addition of purified ABC-F proteins to anin vitrotranslation assay prompts dose-dependent rescue of translation, and demonstrate that such proteins are capable of displacing antibiotic from the ribosomein vitro To our knowledge, these experiments constitute the first direct evidence that ABC-F proteins mediate antibiotic resistance through ribosomal protection.IMPORTANCEAntimicrobial resistance ranks among the greatest threats currently facing human health. Elucidation of the mechanisms by which microorganisms resist the effect of antibiotics is central to understanding the biology of this phenomenon and has the potential to inform the development of new drugs capable of blocking or circumventing resistance. Members of the ABC-F family, which includelsa(A),msr(A),optr(A), andvga(A), collectively yield resistance to a broader range of clinically significant antibiotic classes than any other family of resistance determinants, although their mechanism of action has been controversial since their discovery 25 years ago. Here we present the first direct evidence that proteins of the ABC-F family act to protect the bacterial ribosome from antibiotic-mediated inhibition.
Copyright © 2016 Sharkey et al.
Figures

Comment in
-
The ABC of Ribosome-Related Antibiotic Resistance.mBio. 2016 May 3;7(3):e00598-16. doi: 10.1128/mBio.00598-16. mBio. 2016. PMID: 27143393 Free PMC article.
References
-
- WHO 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland.
-
- Review on Antimicrobial Resistance 2014. Antimicrobial resistance: tackling a crisis for the future health and wealth of nations. Review on Antimicrobial Resistance, London, United Kingdom: http://amr-review.org/Publications.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
