Targeting the epigenetic readers in Ewing sarcoma inhibits the oncogenic transcription factor EWS/Fli1
- PMID: 27006472
- PMCID: PMC5029689
- DOI: 10.18632/oncotarget.8214
Targeting the epigenetic readers in Ewing sarcoma inhibits the oncogenic transcription factor EWS/Fli1
Abstract
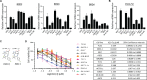
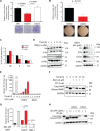
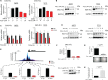
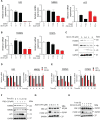
Ewing Sarcoma is a rare bone and soft tissue malignancy affecting children and young adults. Chromosomal translocations in this cancer produce fusion oncogenes as characteristic molecular signatures of the disease. The most common case is the translocation t (11; 22) (q24;q12) which yields the EWS-Fli1 chimeric transcription factor. Finding a way to directly target EWS-Fli1 remains a central therapeutic approach to eradicate this aggressive cancer. Here we demonstrate that treating Ewing Sarcoma cells with JQ1(+), a BET bromodomain inhibitor, represses directly EWS-Fli1 transcription as well as its transcriptional program. Moreover, the Chromatin Immuno Precipitation experiments demonstrate for the first time that these results are a consequence of the depletion of BRD4, one of the BET bromodomains protein from the EWS-Fli1 promoter. In vitro, JQ1(+) treatment reduces the cell viability, impairs the cell clonogenic and the migratory abilities, and induces a G1-phase blockage as well as a time- and a dose-dependent apoptosis. Furthermore, in our in vivo model, we observed a tumor burden delay, an inhibition of the global vascularization and an increase of the mice overall survival. Taken together, our data indicate that inhibiting the BET bromodomains interferes with EWS-FLi1 transcription and could be a promising strategy in the Ewing tumors context.
Keywords: EWS/Fli1; Ewing sarcoma; JQ1; bromodomain; epigenetic.
Conflict of interest statement
Dr. Bradner is the scientific founder of Tensha Therapeutics which has licensed drug like inhibitors of BET bromodomains from the Dana-Farber Cancer Institute for clinical translation as cancer therapeutics.
Figures

Similar articles
-
BET bromodomain inhibitors suppress EWS-FLI1-dependent transcription and the IGF1 autocrine mechanism in Ewing sarcoma.Oncotarget. 2016 Jul 12;7(28):43504-43517. doi: 10.18632/oncotarget.9762. Oncotarget. 2016. PMID: 27259270 Free PMC article.
-
Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma.Oncotarget. 2016 Jan 12;7(2):1451-63. doi: 10.18632/oncotarget.6385. Oncotarget. 2016. PMID: 26623725 Free PMC article.
-
Pharmaceutical Interference of the EWS-FLI1-driven Transcriptome By Cotargeting H3K27ac and RNA Polymerase Activity in Ewing Sarcoma.Mol Cancer Ther. 2021 Oct;20(10):1868-1879. doi: 10.1158/1535-7163.MCT-20-0489. Epub 2021 Jul 26. Mol Cancer Ther. 2021. PMID: 34315769
-
Therapeutic opportunities in Ewing sarcoma: EWS-FLI inhibition via LSD1 targeting.Oncotarget. 2016 Apr 5;7(14):17616-30. doi: 10.18632/oncotarget.7124. Oncotarget. 2016. PMID: 26848860 Free PMC article. Review.
-
Blocking the road, stopping the engine or killing the driver? Advances in targeting EWS/FLI-1 fusion in Ewing sarcoma as novel therapy.Expert Opin Ther Targets. 2014 Nov;18(11):1315-28. doi: 10.1517/14728222.2014.947963. Epub 2014 Aug 27. Expert Opin Ther Targets. 2014. PMID: 25162919 Review.
Cited by
-
AT-hook DNA-binding motif-containing protein one knockdown downregulates EWS-FLI1 transcriptional activity in Ewing's sarcoma cells.PLoS One. 2022 Oct 4;17(10):e0269077. doi: 10.1371/journal.pone.0269077. eCollection 2022. PLoS One. 2022. PMID: 36194562 Free PMC article.
-
Insight into the Etiology of Undifferentiated Soft Tissue Sarcomas from a Novel Mouse Model.Mol Cancer Res. 2019 May;17(5):1024-1035. doi: 10.1158/1541-7786.MCR-18-0117. Epub 2019 Jan 25. Mol Cancer Res. 2019. PMID: 30683671 Free PMC article.
-
One oncogene, several vulnerabilities: EWS/FLI targeted therapies for Ewing sarcoma.J Bone Oncol. 2021 Dec 1;31:100404. doi: 10.1016/j.jbo.2021.100404. eCollection 2021 Dec. J Bone Oncol. 2021. PMID: 34976713 Free PMC article.
-
Immunotherapeutic Challenges for Pediatric Cancers.Mol Ther Oncolytics. 2019 Aug 28;15:38-48. doi: 10.1016/j.omto.2019.08.005. eCollection 2019 Dec 20. Mol Ther Oncolytics. 2019. PMID: 31650024 Free PMC article. Review.
-
Targeting the IGF1R/PI3K/AKT Pathway Sensitizes Ewing Sarcoma to BET Bromodomain Inhibitors.Mol Cancer Ther. 2019 May;18(5):929-936. doi: 10.1158/1535-7163.MCT-18-1151. Epub 2019 Mar 29. Mol Cancer Ther. 2019. PMID: 30926641 Free PMC article.
References
-
- Bernstein M, Kovar H, Paulussen M, Randall RL, Schuck A, Teot LA, Juergens H. Ewing's sarcoma family of tumors: current management. Oncologist. 2006;11:503–519. - PubMed
-
- Ewing J, Classics in oncology. Diffuse endothelioma of bone. James Ewing. Proceedings of the New York Pathological Society, 1921. CA Cancer J Clin. 1972;22:95–98. - PubMed
-
- Schuck A, Ahrens S, Paulussen M, Kuhlen M, Konemann S, Rube C, Winkelmann W, Kotz R, Dunst J, Willich N, Jurgens H. Local therapy in localized Ewing tumors: results of 1058 patients treated in the CESS 81, CESS 86, and EICESS 92 trials. Int J Radiat Oncol Biol Phys. 2003;55:168–177. - PubMed
-
- Grier HE, Krailo MD, Tarbell NJ, Link MP, Fryer CJ, Pritchard DJ, Gebhardt MC, Dickman PS, Perlman EJ, Meyers PA, Donaldson SS, Moore S, Rausen AR, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing's sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003;348:694–701. - PubMed
-
- Paulussen M, Craft AW, Lewis I, Hackshaw A, Douglas C, Dunst J, Schuck A, Winkelmann W, Kohler G, Poremba C, Zoubek A, Ladenstein R, van den Berg H, et al. Results of the EICESS-92 Study: two randomized trials of Ewing's sarcoma treatment—cyclophosphamide compared with ifosfamide in standard-risk patients and assessment of benefit of etoposide added to standard treatment in high-risk patients. J Clin Oncol. 2008;26:4385–4393. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

