Putative oncogene Brachyury (T) is essential to specify cell fate but dispensable for notochord progenitor proliferation and EMT
- PMID: 27006501
- PMCID: PMC4833266
- DOI: 10.1073/pnas.1601252113
Putative oncogene Brachyury (T) is essential to specify cell fate but dispensable for notochord progenitor proliferation and EMT
Abstract
The transcription factor Brachyury (T) gene is expressed throughout primary mesoderm (primitive streak and notochord) during early embryonic development and has been strongly implicated in the genesis of chordoma, a sarcoma of notochord cell origin. Additionally, T expression has been found in and proposed to play a role in promoting epithelial-mesenchymal transition (EMT) in various other types of human tumors. However, the role of T in normal mammalian notochord development and function is still not well-understood. We have generated an inducible knockdown model to efficiently and selectively deplete T from notochord in mouse embryos. In combination with genetic lineage tracing, we show that T function is essential for maintaining notochord cell fate and function. Progenitors adopt predominantly a neural fate in the absence of T, consistent with an origin from a common chordoneural progenitor. However, T function is dispensable for progenitor cell survival, proliferation, and EMT, which has implications for the therapeutic targeting of T in chordoma and other cancers.
Keywords: Brachyury; EMT; cell fate; notochord.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




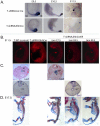

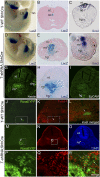

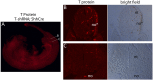

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

