De novo PMP2 mutations in families with type 1 Charcot-Marie-Tooth disease
- PMID: 27009151
- PMCID: PMC5022672
- DOI: 10.1093/brain/aww055
De novo PMP2 mutations in families with type 1 Charcot-Marie-Tooth disease
Abstract
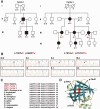
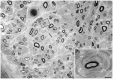
We performed whole exome sequencing on a patient with Charcot-Marie-Tooth disease type 1 and identified a de novo mutation in PMP2, the gene that encodes the myelin P2 protein. This mutation (p.Ile52Thr) was passed from the proband to his one affected son, and segregates with clinical and electrophysiological evidence of demyelinating neuropathy. We then screened a cohort of 136 European probands with uncharacterized genetic cause of Charcot-Marie-Tooth disease and identified another family with Charcot-Marie-Tooth disease type 1 that has a mutation affecting an adjacent amino acid (p.Thr51Pro), which segregates with disease. Our genetic and clinical findings in these kindred demonstrate that dominant PMP2 mutations cause Charcot-Marie-Tooth disease type 1.
Keywords: CMT; Charcot-Marie-Tooth disease; PMP2; myelin P2 protein; peripheral neuropathy.
© The Author (2016). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Barrette B, Calvo E, Vallières N, Lacroix S. Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav Immun 2010; 24: 1254–67. - PubMed
-
- DeArmond SJ, Deibler GE, Bacon M, Kies MW, Eng LF. A neurochemical and immunocytochemical study of P2 protein in human and bovine nervous systems. J Histochem Cytochem 1980; 28: 1275–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

