Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces
- PMID: 27009166
- PMCID: PMC5384728
- DOI: 10.1158/0008-5472.CAN-15-2962
Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces
Abstract
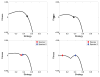
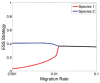
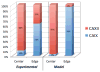
Spatial heterogeneity in tumors is generally thought to result from branching clonal evolution driven by random mutations that accumulate during tumor development. However, this concept rests on the implicit assumption that cancer cells never evolve to a fitness maximum because they can always acquire mutations that increase proliferative capacity. In this study, we investigated the validity of this assumption. Using evolutionary game theory, we demonstrate that local cancer cell populations will rapidly converge to the fittest phenotype given a stable environment. In such settings, cellular spatial heterogeneity in a tumor will be largely governed by regional variations in environmental conditions, for example, alterations in blood flow. Model simulations specifically predict a common spatial pattern in which cancer cells at the tumor-host interface exhibit invasion-promoting, rapidly proliferating phenotypic properties, whereas cells in the tumor core maximize their population density by promoting supportive tissue infrastructures, for example, to promote angiogenesis. We tested model predictions through detailed quantitative image analysis of phenotypic spatial distribution in histologic sections of 10 patients with stage 2 invasive breast cancers. CAIX, GLUT1, and Ki67 were upregulated in the tumor edge, consistent with an acid-producing invasive, proliferative phenotype. Cells in the tumor core were 20% denser than the edge, exhibiting upregulation of CAXII, HIF-1α, and cleaved caspase-3, consistent with a more static and less proliferative phenotype. Similarly, vascularity was consistently lower in the tumor center compared with the tumor edges. Lymphocytic immune responses to tumor antigens also trended to higher level in the tumor edge, although this effect did not reach statistical significance. Like invasive species in nature, cancer cells at the leading edge of the tumor possess a different phenotype from cells in the tumor core. Our results suggest that at least some of the molecular heterogeneity in cancer cells in tumors is governed by predictable regional variations in environmental selection forces, arguing against the assumption that cancer cells can evolve toward a local fitness maximum by random accumulation of mutations. Cancer Res; 76(11); 3136-44. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
Figures
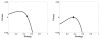

References
-
- Longo DL. Tumor heterogeneity and personalized medicine. N Engl J Med. 2012;366:956–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

