Primitive Embryonic Macrophages are Required for Coronary Development and Maturation
- PMID: 27009605
- PMCID: PMC5567774
- DOI: 10.1161/CIRCRESAHA.115.308270
Primitive Embryonic Macrophages are Required for Coronary Development and Maturation
Abstract
Rationale: It is now recognized that macrophages residing within developing and adult tissues are derived from diverse progenitors including those of embryonic origin. Although the functions of macrophages in adult organisms are well studied, the functions of macrophages during organ development remain largely undefined. Moreover, it is unclear whether distinct macrophage lineages have differing functions.
Objective: To address these issues, we investigated the functions of macrophage subsets resident within the developing heart, an organ replete with embryonic-derived macrophages.
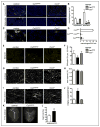
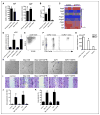
Methods and results: Using a combination of flow cytometry, immunostaining, and genetic lineage tracing, we demonstrate that the developing heart contains a complex array of embryonic macrophage subsets that can be divided into chemokine (C-C motif) receptor 2(-) and chemokine (C-C motif) receptor 2(+) macrophages derived from primitive yolk sac, recombination activating gene 1(+) lymphomyeloid, and Fms-like tyrosine kinase 3(+) fetal monocyte lineages. Functionally, yolk sac-derived chemokine (C-C motif) receptor 2(-) macrophages are instrumental in coronary development where they are required for remodeling of the primitive coronary plexus. Mechanistically, chemokine (C-C motif) receptor 2(-) macrophages are recruited to coronary blood vessels at the onset of coronary perfusion where they mediate coronary plexus remodeling through selective expansion of perfused vasculature. We further demonstrate that insulin like growth factor signaling may mediate the proangiogenic properties of embryonic-derived macrophages.
Conclusions: Together, these findings demonstrate that the embryonic heart contains distinct lineages of embryonic macrophages with unique functions and reveal a novel mechanism that governs coronary development.
Keywords: flow cytometry; fms-like tyrosine kinase 3; macrophages; monocytes; yolk sac.
© 2016 American Heart Association, Inc.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

