Genetic Diversity on the Human X Chromosome Does Not Support a Strict Pseudoautosomal Boundary
- PMID: 27010023
- PMCID: PMC4858793
- DOI: 10.1534/genetics.114.172692
Genetic Diversity on the Human X Chromosome Does Not Support a Strict Pseudoautosomal Boundary
Abstract
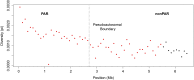
Unlike the autosomes, recombination between the X chromosome and the Y chromosome is often thought to be constrained to two small pseudoautosomal regions (PARs) at the tips of each sex chromosome. PAR1 spans the first 2.7 Mb of the proximal arm of the human sex chromosomes, whereas the much smaller PAR2 encompasses the distal 320 kb of the long arm of each sex chromosome. In addition to PAR1 and PAR2, there is a human-specific X-transposed region that was duplicated from the X to the Y chromosome. The X-transposed region is often not excluded from X-specific analyses, unlike the PARs, because it is not thought to routinely recombine. Genetic diversity is expected to be higher in recombining regions than in nonrecombining regions because recombination reduces the effect of linked selection. In this study, we investigated patterns of genetic diversity in noncoding regions across the entire X chromosome of a global sample of 26 unrelated genetic females. We found that genetic diversity in PAR1 is significantly greater than in the nonrecombining regions (nonPARs). However, rather than an abrupt drop in diversity at the pseudoautosomal boundary, there is a gradual reduction in diversity from the recombining through the nonrecombining regions, suggesting that recombination between the human sex chromosomes spans across the currently defined pseudoautosomal boundary. A consequence of recombination spanning this boundary potentially includes increasing the rate of sex-linked disorders (e.g., de la Chapelle) and sex chromosome aneuploidies. In contrast, diversity in PAR2 is not significantly elevated compared to the nonPARs, suggesting that recombination is not obligatory in PAR2. Finally, diversity in the X-transposed region is higher than in the surrounding nonPARs, providing evidence that recombination may occur with some frequency between the X and Y chromosomes in the X-transposed region.
Keywords: X-transposed region (XTR); genetics of sex; nucleotide diversity; pseudoautosomal region (PAR); recombination; sex chromosome evolution.
Copyright © 2016 by the Genetics Society of America.
Figures


References
-
- Abbas N., McElreavey K., Leconiat M., Vilain E., Jaubert F., et al. , 1993. Familial case of 46,XX male and 46,XX true hermaphrodite associated with a paternal-derived SRY-bearing X chromosome. C. R. Acad. Sci. III 316: 375–383. - PubMed
-
- Bergero R., Charlesworth D., 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24: 94–102. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

