Linking rare and common disease: mapping clinical disease-phenotypes to ontologies in therapeutic target validation
- PMID: 27011785
- PMCID: PMC4804633
- DOI: 10.1186/s13326-016-0051-7
Linking rare and common disease: mapping clinical disease-phenotypes to ontologies in therapeutic target validation
Abstract
Background: The Centre for Therapeutic Target Validation (CTTV - https://www.targetvalidation.org/) was established to generate therapeutic target evidence from genome-scale experiments and analyses. CTTV aims to support the validity of therapeutic targets by integrating existing and newly-generated data. Data integration has been achieved in some resources by mapping metadata such as disease and phenotypes to the Experimental Factor Ontology (EFO). Additionally, the relationship between ontology descriptions of rare and common diseases and their phenotypes can offer insights into shared biological mechanisms and potential drug targets. Ontologies are not ideal for representing the sometimes associated type relationship required. This work addresses two challenges; annotation of diverse big data, and representation of complex, sometimes associated relationships between concepts.
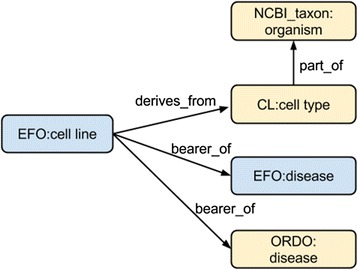
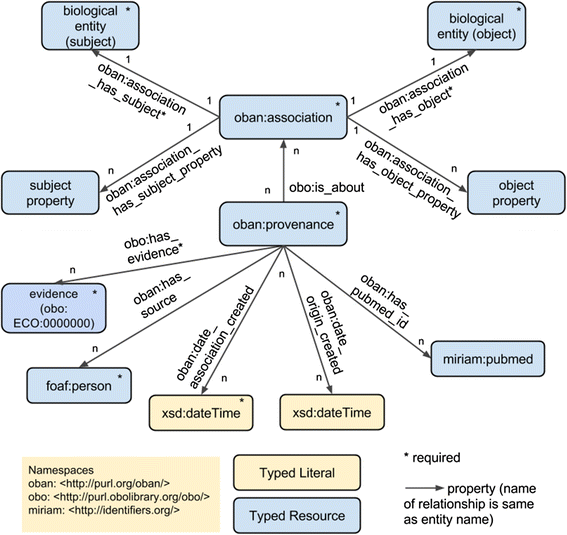
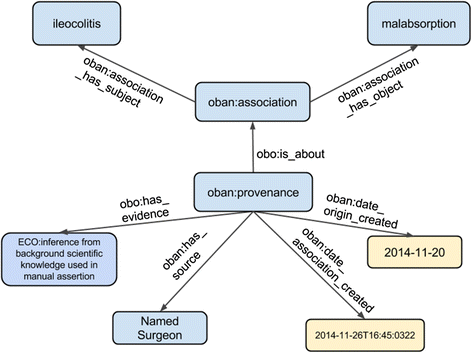
Methods: Semantic mapping uses a combination of custom scripting, our annotation tool 'Zooma', and expert curation. Disease-phenotype associations were generated using literature mining on Europe PubMed Central abstracts, which were manually verified by experts for validity. Representation of the disease-phenotype association was achieved by the Ontology of Biomedical AssociatioN (OBAN), a generic association representation model. OBAN represents associations between a subject and object i.e., disease and its associated phenotypes and the source of evidence for that association. The indirect disease-to-disease associations are exposed through shared phenotypes. This was applied to the use case of linking rare to common diseases at the CTTV.
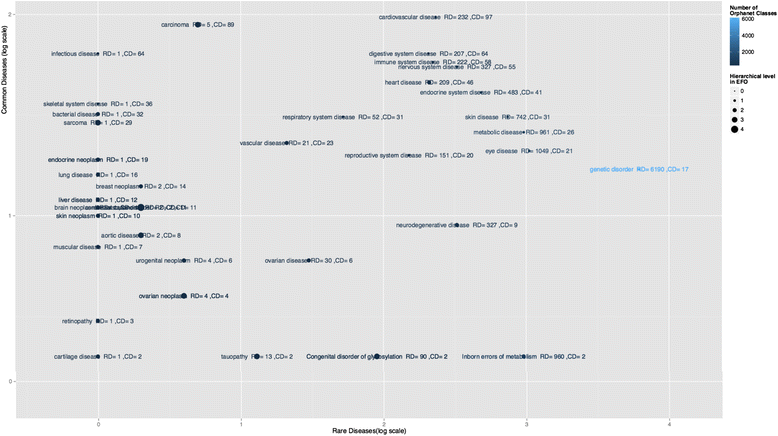
Results: EFO yields an average of over 80% of mapping coverage in all data sources. A 42% precision is obtained from the manual verification of the text-mined disease-phenotype associations. This results in 1452 and 2810 disease-phenotype pairs for IBD and autoimmune disease and contributes towards 11,338 rare diseases associations (merged with existing published work [Am J Hum Genet 97:111-24, 2015]). An OBAN result file is downloadable at http://sourceforge.net/p/efo/code/HEAD/tree/trunk/src/efoassociations/. Twenty common diseases are linked to 85 rare diseases by shared phenotypes. A generalizable OBAN model for association representation is presented in this study.
Conclusions: Here we present solutions to large-scale annotation-ontology mapping in the CTTV knowledge base, a process for disease-phenotype mining, and propose a generic association model, 'OBAN', as a means to integrate disease using shared phenotypes.
Availability: EFO is released monthly and available for download at http://www.ebi.ac.uk/efo/.
Keywords: CTTV; EFO; OBAN; Phenotype disease associations; Rare disease.
Figures


References
-
- McKusick-Nathans Institute of Genetic Medicine JHU. Online Mendelian Inheritance in Man, OMIM. Baltimore, MD. 1998. http://www.omim.org/. 2015.
-
- INSERM-Orphanet. Orphanet: an online database of rare diseases and orphan drugs. Paris, France. 1997. http://www.orpha.net/. 2015.
-
- Vasant D, Chanas L, Malone J, Hanauer M, Olry A, Jupp S, et al. ORDO: An Ontology Connecting Rare Disease, Epidemiology and Genetic Data. 2014.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

