Ubiquitin modifications
- PMID: 27012465
- PMCID: PMC4822133
- DOI: 10.1038/cr.2016.39
Ubiquitin modifications
Abstract
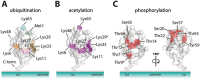
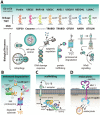
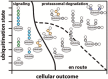
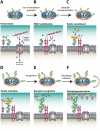
Protein ubiquitination is a dynamic multifaceted post-translational modification involved in nearly all aspects of eukaryotic biology. Once attached to a substrate, the 76-amino acid protein ubiquitin is subjected to further modifications, creating a multitude of distinct signals with distinct cellular outcomes, referred to as the 'ubiquitin code'. Ubiquitin can be ubiquitinated on seven lysine (Lys) residues or on the N-terminus, leading to polyubiquitin chains that can encompass complex topologies. Alternatively or in addition, ubiquitin Lys residues can be modified by ubiquitin-like molecules (such as SUMO or NEDD8). Finally, ubiquitin can also be acetylated on Lys, or phosphorylated on Ser, Thr or Tyr residues, and each modification has the potential to dramatically alter the signaling outcome. While the number of distinctly modified ubiquitin species in cells is mind-boggling, much progress has been made to characterize the roles of distinct ubiquitin modifications, and many enzymes and receptors have been identified that create, recognize or remove these ubiquitin modifications. We here provide an overview of the various ubiquitin modifications present in cells, and highlight recent progress on ubiquitin chain biology. We then discuss the recent findings in the field of ubiquitin acetylation and phosphorylation, with a focus on Ser65-phosphorylation and its role in mitophagy and Parkin activation.
Figures




References
-
- Cohen P. The origins of protein phosphorylation. Nat Cell Biol 2002; 4:E127–E130. - PubMed
-
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol 2015; 16:258–264. - PubMed
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425–479. - PubMed
-
- Clague MJ, Heride C, Urbé S. The demographics of the ubiquitin system. Trends Cell Biol 2015; 25:417–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

