The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement
- PMID: 27013146
- PMCID: PMC4888456
- DOI: 10.1042/BCJ20160245
The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement
Abstract
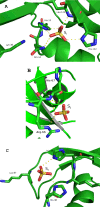
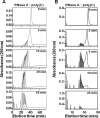
Human RNase 6 is a cationic secreted protein that belongs to the RNase A superfamily. Its expression is induced in neutrophils and monocytes upon bacterial infection, suggesting a role in host defence. We present here the crystal structure of RNase 6 obtained at 1.72 Å (1 Å=0.1 nm) resolution, which is the first report for the protein 3D structure and thereby setting the basis for functional studies. The structure shows an overall kidney-shaped globular fold shared with the other known family members. Three sulfate anions bound to RNase 6 were found, interacting with residues at the main active site (His(15), His(122) and Gln(14)) and cationic surface-exposed residues (His(36), His(39), Arg(66) and His(67)). Kinetic characterization, together with prediction of protein-nucleotide complexes by molecular dynamics, was applied to analyse the RNase 6 substrate nitrogenous base and phosphate selectivity. Our results reveal that, although RNase 6 is a moderate catalyst in comparison with the pancreatic RNase type, its structure includes lineage-specific features that facilitate its activity towards polymeric nucleotide substrates. In particular, enzyme interactions at the substrate 5' end can provide an endonuclease-type cleavage pattern. Interestingly, the RNase 6 crystal structure revealed a novel secondary active site conformed by the His(36)-His(39) dyad that facilitates the polynucleotide substrate catalysis.
Keywords: RNase A superfamily; RNase k6; kinetic characterization; molecular dynamics; protein crystallography; sulfate anion.
© 2016 The Author(s). published by Portland Press Limited on behalf of the Biochemical Society.
Figures





Similar articles
-
Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.Biochim Biophys Acta Gen Subj. 2019 Jan;1863(1):105-117. doi: 10.1016/j.bbagen.2018.09.021. Epub 2018 Oct 1. Biochim Biophys Acta Gen Subj. 2019. PMID: 30287244
-
Structural basis of substrate specificity in porcine RNase 4.FEBS J. 2016 Mar;283(5):912-28. doi: 10.1111/febs.13646. Epub 2016 Feb 5. FEBS J. 2016. PMID: 26748441
-
Crystal structure of eosinophil cationic protein at 2.4 A resolution.Biochemistry. 1999 Dec 21;38(51):16794-801. doi: 10.1021/bi9919145. Biochemistry. 1999. PMID: 10606511
-
Structural and functional importance of local and global conformational fluctuations in the RNase A superfamily.FEBS J. 2013 Nov;280(22):5596-607. doi: 10.1111/febs.12371. Epub 2013 Jun 24. FEBS J. 2013. PMID: 23763751 Review.
-
Bovine pancreatic ribonuclease A as a model of an enzyme with multiple substrate binding sites.Biochim Biophys Acta. 1995 Nov 15;1253(1):16-24. doi: 10.1016/0167-4838(95)00138-k. Biochim Biophys Acta. 1995. PMID: 7492594 Review.
Cited by
-
Nucleotide modifications enable rational design of TLR7-selective ligands by blocking RNase cleavage.J Exp Med. 2024 Feb 5;221(2):e20230341. doi: 10.1084/jem.20230341. Epub 2023 Dec 14. J Exp Med. 2024. PMID: 38095631 Free PMC article.
-
Biological Activities of Secretory RNases: Focus on Their Oligomerization to Design Antitumor Drugs.Front Immunol. 2019 Nov 26;10:2626. doi: 10.3389/fimmu.2019.02626. eCollection 2019. Front Immunol. 2019. PMID: 31849926 Free PMC article. Review.
-
A Common Polymorphism in RNASE6 Impacts Its Antimicrobial Activity toward Uropathogenic Escherichia coli.Int J Mol Sci. 2024 Jan 3;25(1):604. doi: 10.3390/ijms25010604. Int J Mol Sci. 2024. PMID: 38203775 Free PMC article.
-
Insights into the Antimicrobial Mechanism of Action of Human RNase6: Structural Determinants for Bacterial Cell Agglutination and Membrane Permeation.Int J Mol Sci. 2016 Apr 13;17(4):552. doi: 10.3390/ijms17040552. Int J Mol Sci. 2016. PMID: 27089320 Free PMC article.
-
Evolutionary Trends in RNA Base Selectivity Within the RNase A Superfamily.Front Pharmacol. 2019 Oct 9;10:1170. doi: 10.3389/fphar.2019.01170. eCollection 2019. Front Pharmacol. 2019. PMID: 31649540 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

